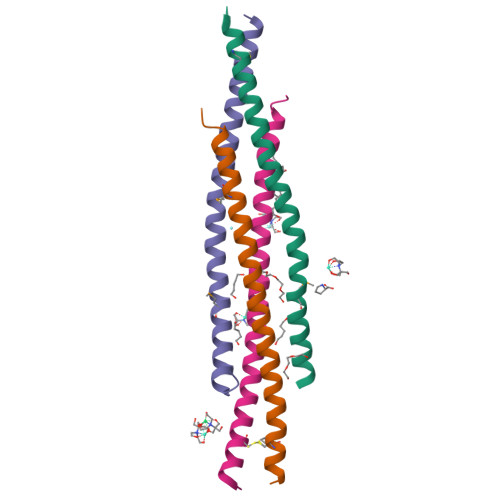
The structure of a NEMO construct engineered for screening reveals novel determinants of inhibition.
Kennedy, A.E., Barczewski, A.H., Arnoldy, C.R., Pennington, J.P., Tiernan, K.A., Hidalgo, M.B., Reilly, C.C., Wongsri, T., Ragusa, M.J., Grigoryan, G., Mierke, D.F., Pellegrini, M.(2025) Structure
- PubMed: 39909030
- DOI: https://doi.org/10.1016/j.str.2025.01.010
- Primary Citation of Related Structures:
8U7C - PubMed Abstract:
NEMO is an essential component in the activation of the canonical nuclear factor κB (NF-κB) pathway and exerts its function by recruiting the IκB kinases (IKK) to the IKK complex. Inhibition of the NEMO/IKKs interaction is an attractive therapeutic paradigm for diseases related to NF-κB mis-regulation, but a difficult endeavor because of the extensive protein-protein interface. Here we report the design and characterization of novel engineered constructs of the IKK-binding domain of NEMO, programmed to render this difficult protein domain amenable to NMR measurements and crystallization, while preserving its biological function. ZipNEMO binds IKKβ with nanomolar affinity, is amenable to heteronuclear nuclear magnetic resonance (NMR) techniques and structure determination by X-ray crystallography. We show that NMR spectra of zipNEMO allow to detect inhibitor binding in solution and resonance assignment. The crystal structure of zipNEMO reveals a novel ligand binding motif and the adaptability of the binding pocket and inspired the design of new peptide inhibitors.
Organizational Affiliation:
Department of Chemistry, Dartmouth College, Hanover, NH 03755, USA.