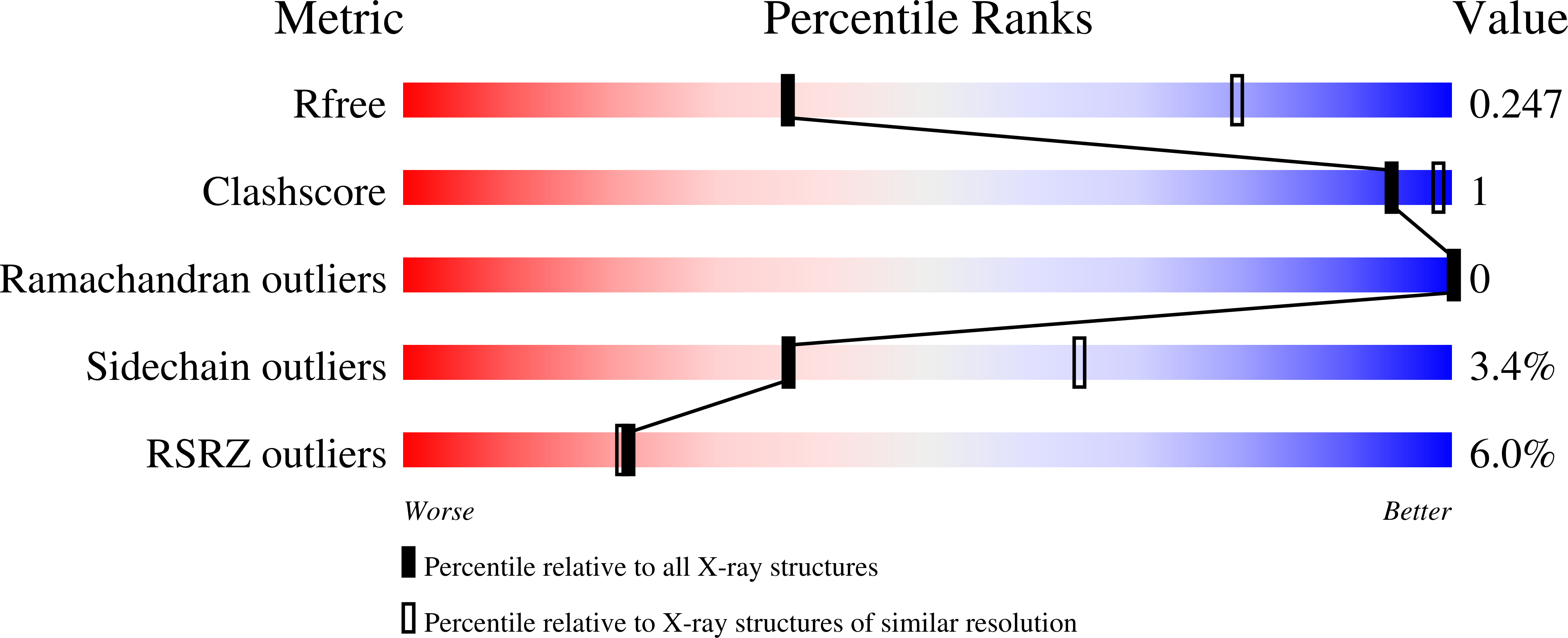
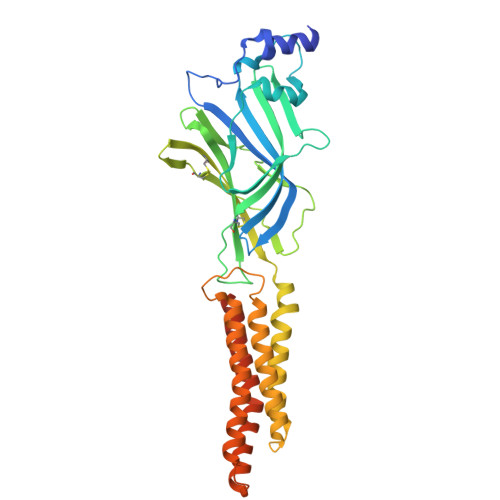
Crystal Structures of Human GlyRa3 Bound to a Novel Class of Potentiators with Efficacy in a Mouse Model of Neuropathic Pain
Huang, X., Shaffer, P.L., Ayube, S., Bregman, H., Chen, H., Lehto, S.G., Luther, J.A., Matson, D.J., McDonough, S.I., Michelsen, K., Plant, M.M., Schneider, S., Simard, J.R., Teffera, Y., Yi, S., Zhang, M., DiMauro, E.F., Gingras, J.To be published.