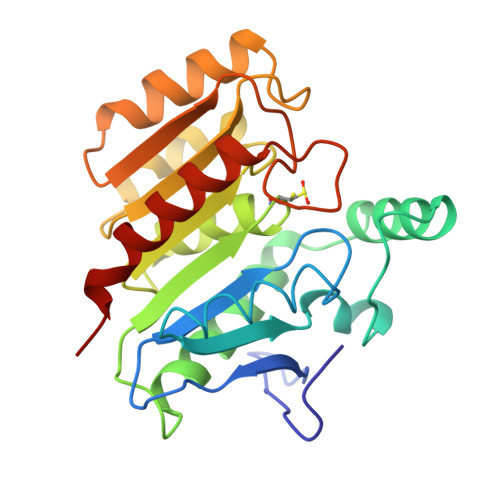
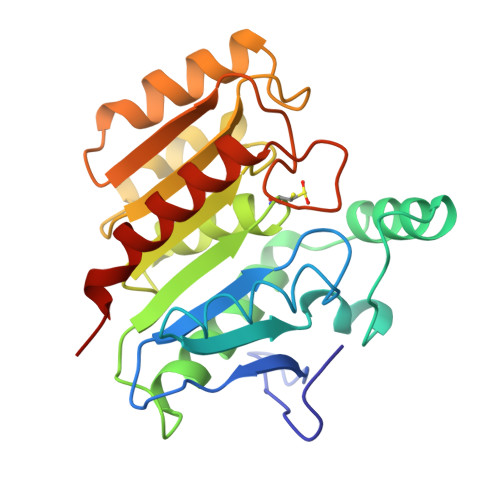
Refined structure of dienelactone hydrolase at 1.8 A.
Pathak, D., Ollis, D.(1990) J Mol Biology 214: 497-525
- PubMed: 2380986
- DOI: https://doi.org/10.1016/0022-2836(90)90196-s
- Primary Citation of Related Structures:
1DIN - PubMed Abstract:
The structure of dienelactone hydrolase (DLH) from Pseudomonus sp. B13, after stereochemically restrained least-squares refinement at 1.8 A resolution, is described. The final molecular model of DLH has a conventional R value of 0.150 and includes all but the carboxyl-terminal three residues that are crystallographically disordered. The positions of 279 water molecules are included in the final model. The root-mean-square deviation from ideal bond distances for the model is 0.014 A and the error in atomic co-ordinates is estimated to be 0.15 A. DLH is a monomeric enzyme containing 236 amino acid residues and is a member of the beta-ketoadipate pathway found in bacteria and fungi. DLH is an alpha/beta protein containing seven helices and eight strands of beta-pleated sheet. A single 4-turn 3(10)-helix is seen. The active-site Cys123 residues at the N-terminal end of an alpha-helix that is peculiar in its consisting entirely of hydrophobic residues (except for a C-terminal lysine). The beta-sheet is composed of parallel strands except for strand 2, which gives rise to a short antiparallel region at the N-terminal end of the central beta-sheet. The active-site cysteine residue is part of a triad of residues consisting of Cys123, His202 and Asp171, and is reminiscent of the serine/cysteine proteases. As in papain and actinidin, the active thiol is partially oxidized during X-ray data collection. The positions of both the reduced and the oxidized sulphur are described. The active site geometry suggests that a change in the conformation of the native thiol occurs upon diffusion of substrate into the active site cleft of DLH. This enables nucleophilic attack by the gamma-sulphur to occur on the cyclic ester substrate through a ring-opening reaction.
Organizational Affiliation:
Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, IL 60208.