Structural basis of Integrator-dependent RNA polymerase II termination.
Fianu, I., Ochmann, M., Walshe, J.L., Dybkov, O., Cruz, J.N., Urlaub, H., Cramer, P.(2024) Nature 629: 219-227
- PubMed: 38570683
- DOI: https://doi.org/10.1038/s41586-024-07269-4
- Primary Citation of Related Structures:
8RBX, 8RBZ, 8RC4 - PubMed Abstract:
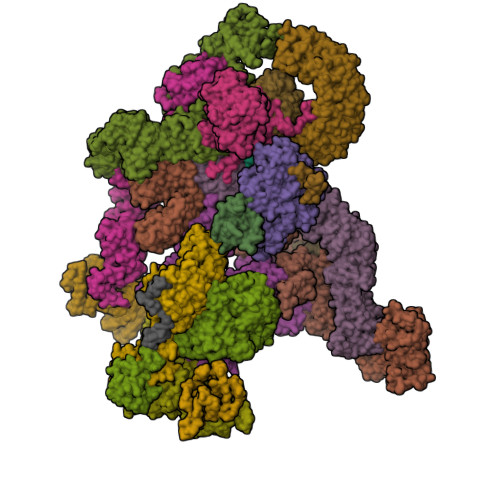
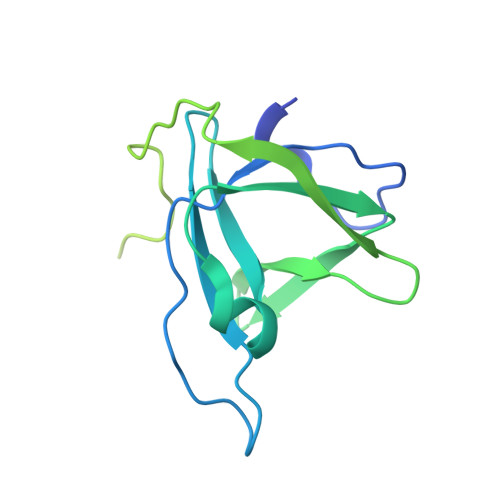
The Integrator complex can terminate RNA polymerase II (Pol II) in the promoter-proximal region of genes. Previous work has shed light on how Integrator binds to the paused elongation complex consisting of Pol II, the DRB sensitivity-inducing factor (DSIF) and the negative elongation factor (NELF) and how it cleaves the nascent RNA transcript 1 , but has not explained how Integrator removes Pol II from the DNA template. Here we present three cryo-electron microscopy structures of the complete Integrator-PP2A complex in different functional states. The structure of the pre-termination complex reveals a previously unresolved, scorpion-tail-shaped INTS10-INTS13-INTS14-INTS15 module that may use its 'sting' to open the DSIF DNA clamp and facilitate termination. The structure of the post-termination complex shows that the previously unresolved subunit INTS3 and associated sensor of single-stranded DNA complex (SOSS) factors prevent Pol II rebinding to Integrator after termination. The structure of the free Integrator-PP2A complex in an inactive closed conformation 2 reveals that INTS6 blocks the PP2A phosphatase active site. These results lead to a model for how Integrator terminates Pol II transcription in three steps that involve major rearrangements.
Organizational Affiliation:
Department of Molecular Biology, Max Planck Institute for Multidisciplinary Sciences, Göttingen, Germany. isaac.fianu@mpinat.mpg.de.