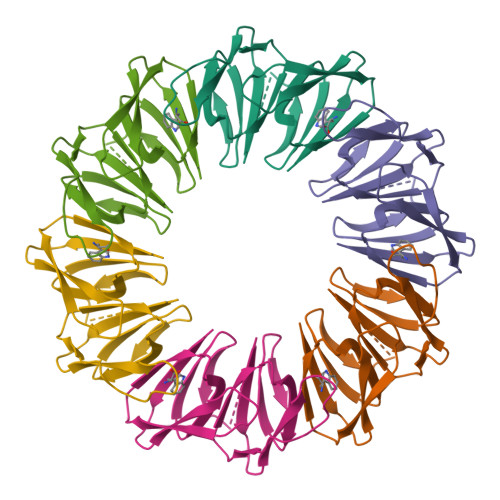
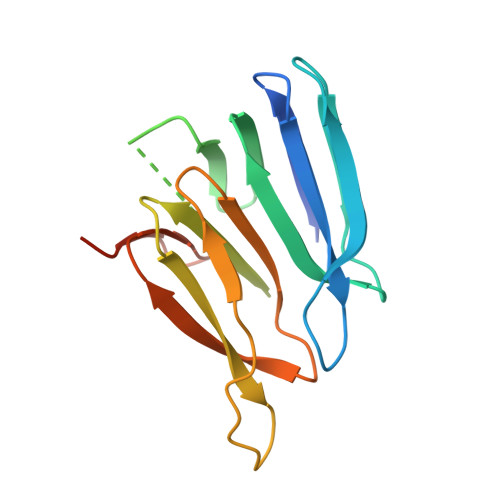
Structural basis of nearest-neighbor cooperativity in the ring-shaped gene regulatory protein TRAP from protein engineering and cryo-EM.
Li, W., Yang, H., Stachowski, K., Norris, A.S., Lichtenthal, K., Kelly, S., Gollnick, P., Wysocki, V.H., Foster, M.P.(2025) Proc Natl Acad Sci U S A 122: e2409030121-e2409030121
- PubMed: 39793047
- DOI: https://doi.org/10.1073/pnas.2409030121
- Primary Citation of Related Structures:
9BDS, 9BE7, 9BE8 - PubMed Abstract:
The homo-dodecameric ring-shaped trp RNA binding attenuation protein (TRAP) from Alkalihalobacillus halodurans (Aha) binds up to twelve tryptophan ligands (Trp) and becomes activated to bind a specific sequence in the 5' leader region of the trp operon mRNA, thereby downregulating biosynthesis of Trp. Thermodynamic measurements of Trp binding have revealed a range of cooperative behavior for different TRAP variants, even if the averaged apparent affinities for Trp have been found to be similar. Proximity between the ligand binding sites, and the ligand-coupled disorder-to-order transition has implicated nearest-neighbor interactions in cooperativity. To establish a solid basis for describing nearest-neighbor cooperativity in TRAP, we engineered variants constructed with two subunits connected by a flexible linker (dTRAP). We mutated the binding sites of alternating protomers such that only every other site was competent for Trp binding (WT-Mut dTRAP). Ligand binding monitored by NMR, calorimetry, and native mass spectrometry revealed strong cooperativity in dTRAP containing adjacent binding-competent sites, but a severe binding defect when the wild-type sites were separated by mutated sites. Cryo-EM experiments of dTRAP in its ligand-free apo state, and both dTRAP and WT-Mut dTRAP in the presence of Trp, revealed progressive stabilization of loops that gate the Trp binding site and participate in RNA binding. These studies provide important insights into the thermodynamic and structural basis for the observed ligand binding cooperativity in TRAP. Such insights can be useful for understanding allosteric control networks and for the development of those with defined ligand sensitivity and regulatory control.
Organizational Affiliation:
Department of Chemistry and Biochemistry, The Ohio State University, Columbus, OH 43210.