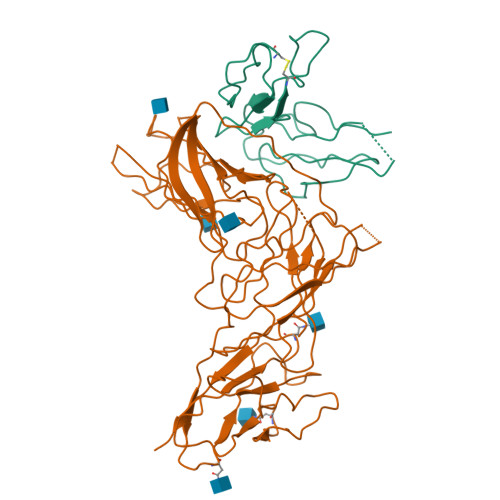
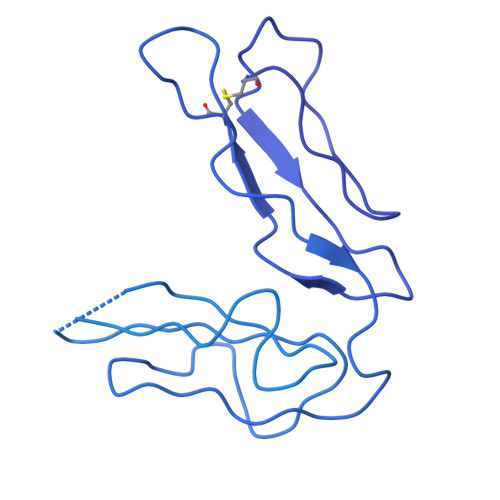
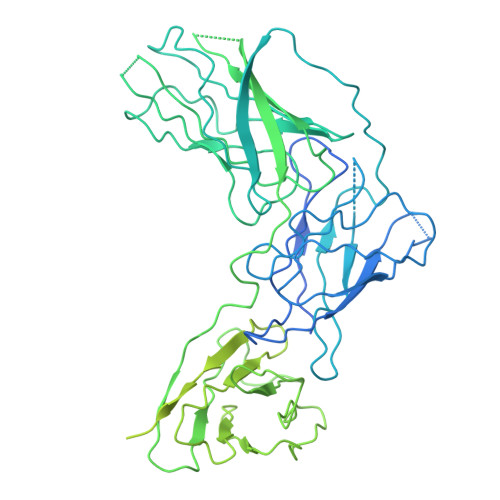
Structural basis of Epstein-Barr virus gp350 receptor recognition and neutralization.
Sun, C., Fang, X.Y., Bu, G.L., Zhong, L.Y., Xie, C., Zhao, G.X., Sui, S.F., Liu, Z., Zeng, M.S.(2025) Cell Rep 44: 115168-115168
- PubMed: 39792550
- DOI: https://doi.org/10.1016/j.celrep.2024.115168
- Primary Citation of Related Structures:
8ZNI - PubMed Abstract:
Epstein-Barr virus (EBV) is an oncogenic virus associated with multiple lymphoid malignancies and autoimmune diseases. During infection in B cells, EBV uses its major glycoprotein gp350 to recognize the host receptor CR2, initiating viral attachment, a process that has lacked direct structural evidence for decades. In this study, we resolved the structure of the gp350-CR2 complex, elucidated their key interactions, and determined the site-specific N-glycosylation map of gp350. Our findings reveal that CR2 primarily binds to gp350 through an electrostatically complementary and glycan-free interface and that the diversity of key residues in CR2 across different species influences EBV host selectivity mediated by gp350. With the confirmed binding, we constructed a CR2-Fc antibody analog that targets the vulnerable site of gp350, demonstrating a potent neutralization effect against EBV infection in B cells. Our work provides essential structural insights into the mechanism of EBV infection and host tropism, suggesting a potential antiviral agent.
Organizational Affiliation:
State Key Laboratory of Oncology in South China, Guangdong Provincial Clinical Research Center for Cancer, Guangdong Key Laboratory of Nasopharyngeal Carcinoma Diagnosis and Therapy, Sun Yat-sen University Cancer Center, Guangzhou 510060, China. Electronic address: suncong@sysucc.org.cn.