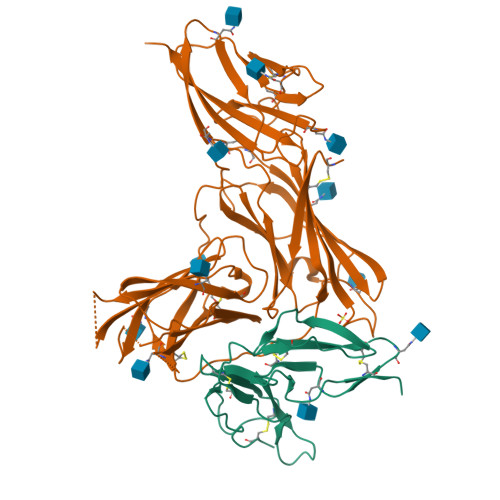
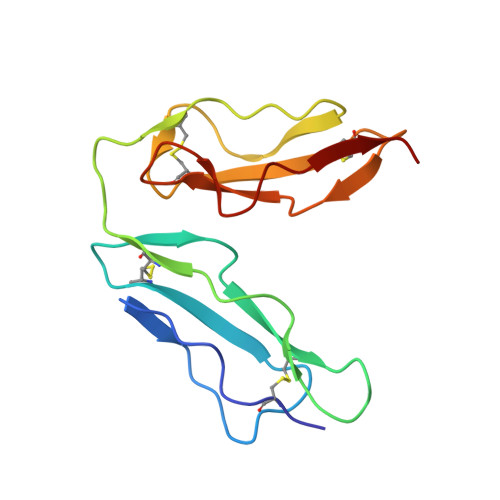
Structural basis for complement receptor engagement and virus neutralization through Epstein-Barr virus gp350.
Joyce, M.G., Bu, W., Chen, W.H., Gillespie, R.A., Andrews, S.F., Wheatley, A.K., Tsybovsky, Y., Jensen, J.L., Stephens, T., Prabhakaran, M., Fisher, B.E., Narpala, S.R., Bagchi, M., McDermott, A.B., Nabel, G.J., Kwong, P.D., Mascola, J.R., Cohen, J.I., Kanekiyo, M.(2025) Immunity 58: 295
- PubMed: 39909035
- DOI: https://doi.org/10.1016/j.immuni.2025.01.010
- Primary Citation of Related Structures:
8SEF, 8SGA, 8SGG, 8SGN, 8SIC, 8SM0, 8SM1 - PubMed Abstract:
Epstein-Barr virus (EBV) causes infectious mononucleosis and is associated with malignancies in humans. Viral infection of B cells is initiated by the viral glycoprotein 350 (gp350) binding to complement receptor 2 (CR2). Despite decades of effort, no vaccines or curative agents have been developed, partly due to lack of atomic-level understanding of the virus-host interface. Here, we determined the 1.7 Å structure of gp350 in complex with CR2. CR2 binding of gp350 utilized the same set of Arg residues required for recognition of its natural ligand, complement C3d. We further determined the structures of gp350 in complex with three potently neutralizing antibodies (nAbs) obtained from vaccinated macaques and EBV-infected individuals. Like the CR2 interaction, these nAbs targeted the acidic pocket within the CR2-binding site on gp350 using Arg residues. Our results illustrate two axes of molecular mimicry-gp350 versus C3d and CR2 versus EBV nAbs-offering insights for EBV vaccines and therapeutics development.
Organizational Affiliation:
Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA; Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc, Bethesda, MD 20817, USA; Emerging Infectious Diseases Branch, Walter Reed Army Institute of Research, Silver Spring, MD 20910, USA. Electronic address: gjoyce@eidresearch.org.