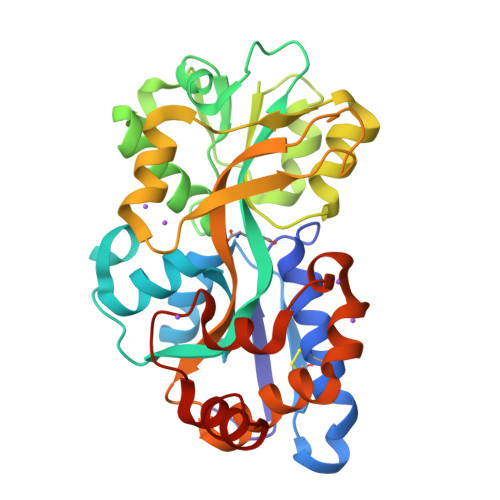
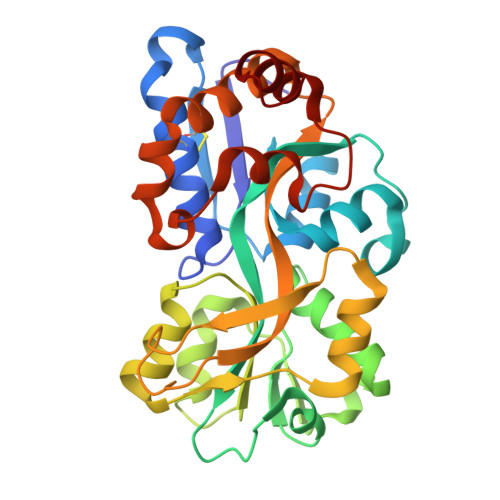
Structure and selectivity of a glutamate-specific TAXI TRAP binding protein from Vibrio cholerae.
Davies, J.F.S., Daab, A., Massouh, N., Kirkland, C., Strongitharm, B., Leech, A., Farre, M., Thomas, G.H., Mulligan, C.(2024) J Gen Physiol 156
- PubMed: 39556531
- DOI: https://doi.org/10.1085/jgp.202413584
- Primary Citation of Related Structures:
8S4J - PubMed Abstract:
Tripartite ATP-independent periplasmic (TRAP) transporters are widespread in prokaryotes and are responsible for the transport of a variety of different ligands, primarily organic acids. TRAP transporters can be divided into two subclasses; DctP-type and TAXI type, which share the same overall architecture and substrate-binding protein requirement. DctP-type transporters are very well studied and have been shown to transport a range of compounds including dicarboxylates, keto acids, and sugar acids. However, TAXI-type transporters are relatively poorly understood. To address this gap in our understanding, we have structurally and biochemically characterized VC0430 from Vibrio cholerae. We show it is a monomeric, high affinity glutamate-binding protein, which we thus rename VcGluP. VcGluP is stereoselective, binding the L-isomer preferentially, and can also bind L-glutamine and L-pyroglutamate with lower affinity. Structural characterization of ligand-bound VcGluP revealed details of its binding site and biophysical characterization of binding site mutants revealed the substrate binding determinants, which differ substantially from those of DctP-type TRAPs. Finally, we have analyzed the interaction between VcGluP and its cognate membrane component, VcGluQM (formerly VC0429) in silico, revealing an architecture hitherto unseen. To our knowledge, this is the first transporter in V. cholerae to be identified as specific to glutamate, which plays a key role in the osmoadaptation of V. cholerae, making this transporter a potential therapeutic target.
Organizational Affiliation:
School of Biosciences, Division of Natural Sciences, University of Kent, Canterbury, UK.