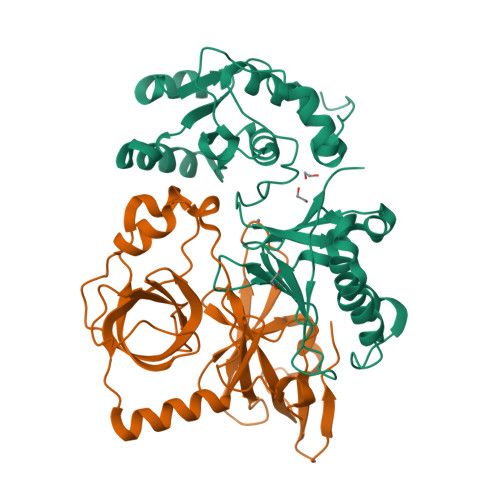
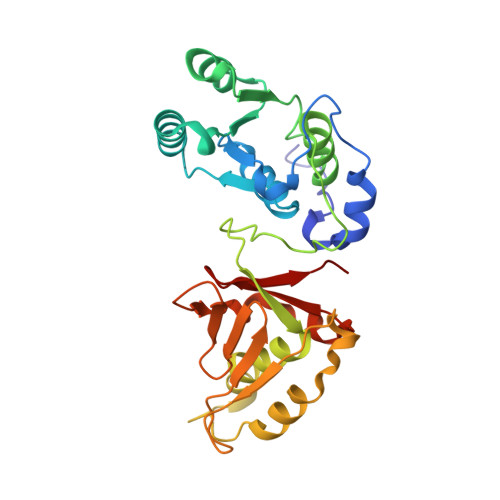
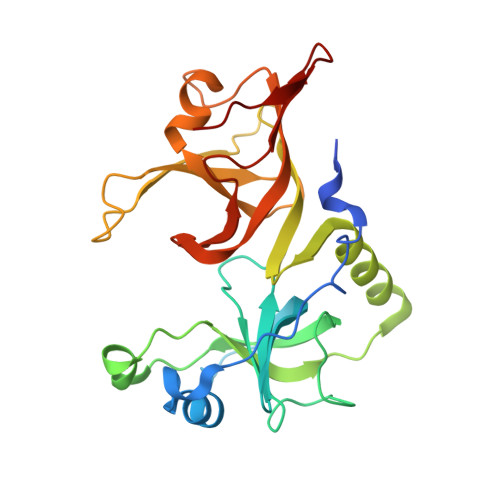
MUT-7 exoribonuclease activity and localization are mediated by an ancient domain.
Busetto, V., Pshanichnaya, L., Lichtenberger, R., Hann, S., Ketting, R.F., Falk, S.(2024) Nucleic Acids Res 52: 9076-9091
- PubMed: 39188014
- DOI: https://doi.org/10.1093/nar/gkae610
- Primary Citation of Related Structures:
8Q66 - PubMed Abstract:
The MUT-7 family of 3'-5' exoribonucleases is evolutionarily conserved across the animal kingdom and plays essential roles in small RNA production in the germline. Most MUT-7 homologues carry a C-terminal domain of unknown function named MUT7-C appended to the exoribonuclease domain. Our analysis shows that the MUT7-C is evolutionary ancient, as a minimal version of the domain exists as an individual protein in prokaryotes. In animals, MUT7-C has acquired an insertion that diverged during evolution, expanding its functions. Caenorhabditis elegans MUT-7 contains a specific insertion within MUT7-C, which allows binding to MUT-8 and, consequently, MUT-7 recruitment to germ granules. In addition, in C. elegans and human MUT-7, the MUT7-C domain contributes to RNA binding and is thereby crucial for ribonuclease activity. This RNA-binding function most likely represents the ancestral function of the MUT7-C domain. Overall, this study sheds light on MUT7-C and assigns two functions to this previously uncharacterized domain.
Organizational Affiliation:
Max Perutz Labs, Vienna Biocenter Campus (VBC), Dr.-Bohr-Gasse 9, 1030 Vienna, Austria.