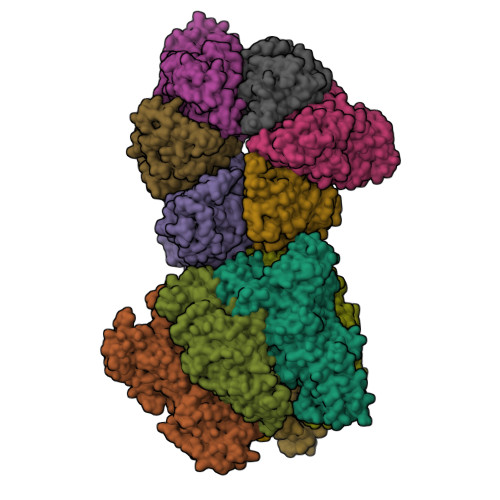
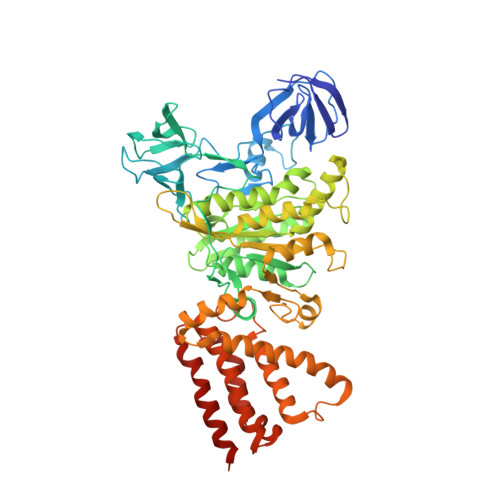
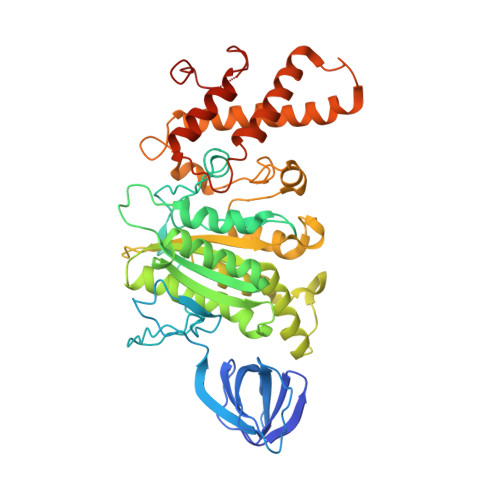
The combination of high-speed AFM and X-ray crystallography reveals rotary catalysis
Imamura, M., Maruyama, S., Nakamoto, K., Kawai, F., Akiyama, T., Mizutani, K., Suzuki, K., Shirouzu, M., Iino, R., Uchihashi, T., Ando, T., Murata, T.To be published.