Endogenous agonist-bound S1PR3 structure reveals determinants of G protein-subtype bias.
Maeda, S., Shiimura, Y., Asada, H., Hirata, K., Luo, F., Nango, E., Tanaka, N., Toyomoto, M., Inoue, A., Aoki, J., Iwata, S., Hagiwara, M.(2021) Sci Adv 7
- PubMed: 34108205
- DOI: https://doi.org/10.1126/sciadv.abf5325
- Primary Citation of Related Structures:
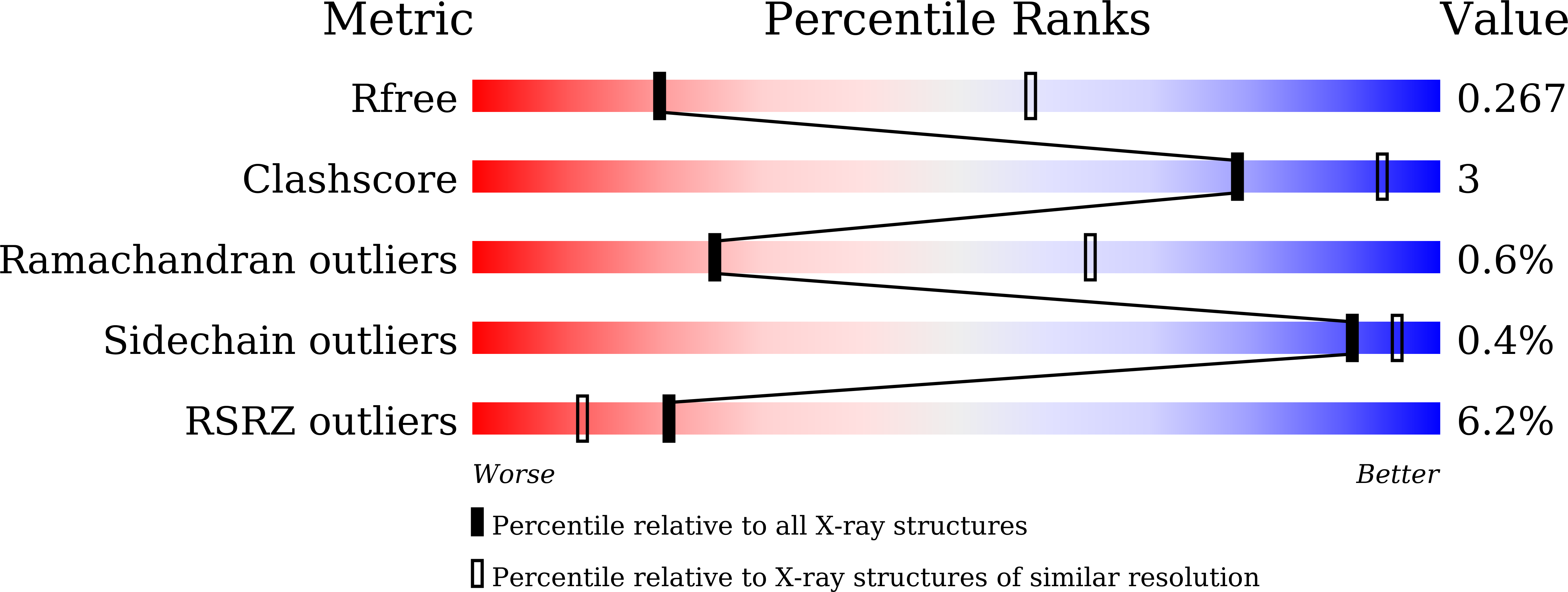
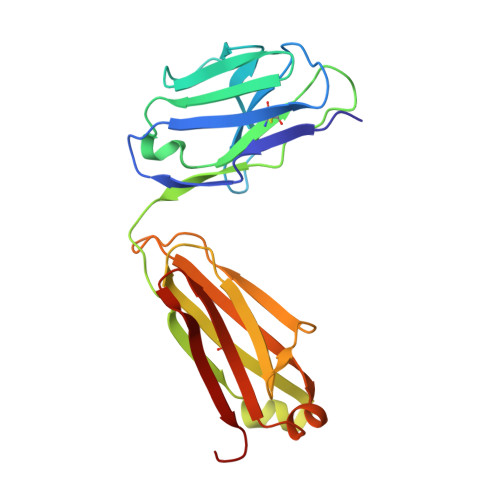
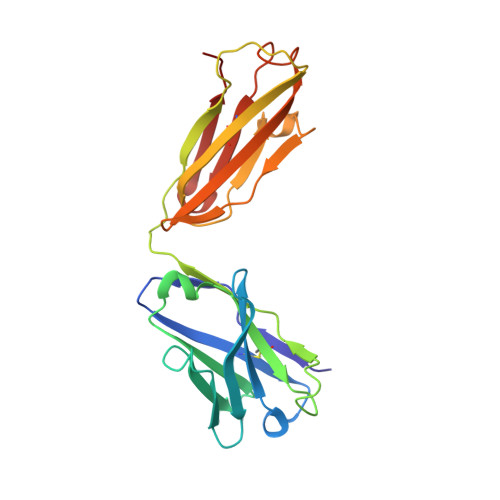
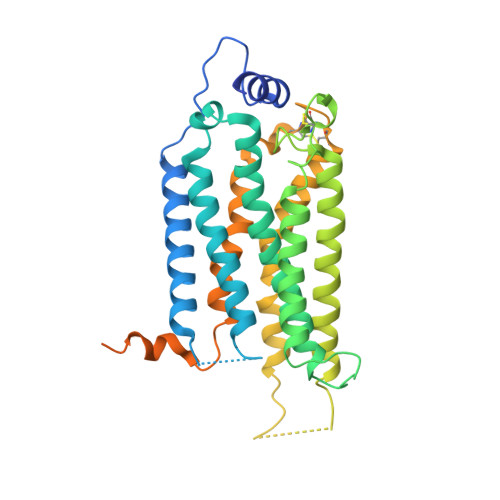
7C4S - PubMed Abstract:
Sphingosine-1-phosphate (S1P) regulates numerous important physiological functions, including immune response and vascular integrity, via its cognate receptors (S1PR1 to S1PR5); however, it remains unclear how S1P activates S1PRs upon binding. Here, we determined the crystal structure of the active human S1PR3 in complex with its natural agonist S1P at 3.2-Å resolution. S1P exhibits an unbent conformation in the long tunnel, which penetrates through the receptor obliquely. Compared with the inactive S1PR1 structure, four residues surrounding the alkyl tail of S1P (the "quartet core") exhibit orchestrating rotamer changes that accommodate the moiety, thereby inducing an active conformation. In addition, we reveal that the quartet core determines G protein selectivity of S1PR3. These results offer insight into the structural basis of activation and biased signaling in G protein-coupled receptors and will help the design of biased ligands for optimized therapeutics.
Organizational Affiliation:
Department of Anatomy and Developmental Biology, Graduate School of Medicine, Kyoto University, Kyoto 606-8501, Japan.