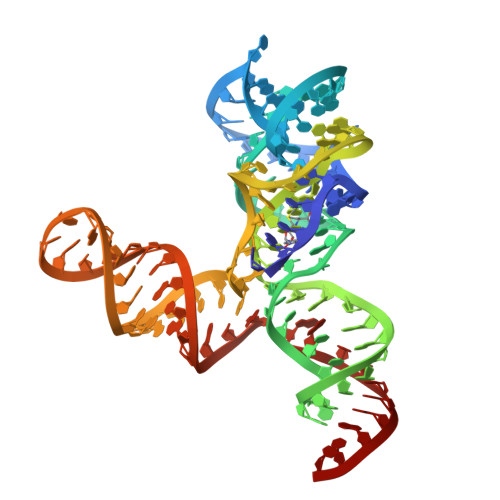
Cryo-EM structure of a 40 kDa SAM-IV riboswitch RNA at 3.7 angstrom resolution.
Zhang, K., Li, S., Kappel, K., Pintilie, G., Su, Z., Mou, T.C., Schmid, M.F., Das, R., Chiu, W.(2019) Nat Commun 10: 5511-5511
- PubMed: 31796736
- DOI: https://doi.org/10.1038/s41467-019-13494-7
- Primary Citation of Related Structures:
6UES, 6UET - PubMed Abstract:
Specimens below 50 kDa have generally been considered too small to be analyzed by single-particle cryo-electron microscopy (cryo-EM). The high flexibility of pure RNAs makes it difficult to obtain high-resolution structures by cryo-EM. In bacteria, riboswitches regulate sulfur metabolism through binding to the S-adenosylmethionine (SAM) ligand and offer compelling targets for new antibiotics. SAM-I, SAM-I/IV, and SAM-IV are the three most commonly found SAM riboswitches, but the structure of SAM-IV is still unknown. Here, we report the structures of apo and SAM-bound SAM-IV riboswitches (119-nt, ~40 kDa) to 3.7 Å and 4.1 Å resolution, respectively, using cryo-EM. The structures illustrate homologies in the ligand-binding core but distinct peripheral tertiary contacts in SAM-IV compared to SAM-I and SAM-I/IV. Our results demonstrate the feasibility of resolving small RNAs with enough detail to enable detection of their ligand-binding pockets and suggest that cryo-EM could play a role in structure-assisted drug design for RNA.
Organizational Affiliation:
Department of Bioengineering, and James H. Clark Center, Stanford University, Stanford, CA, 94305, USA.