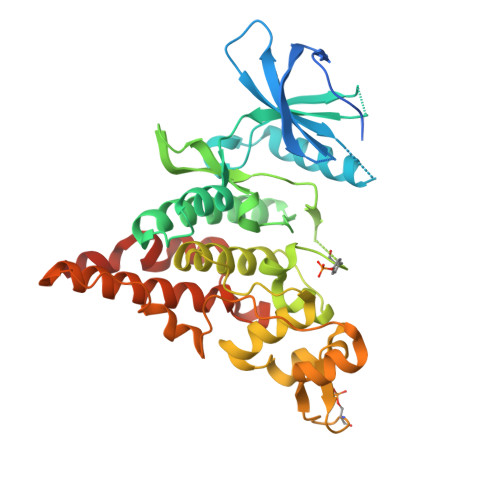
The crystal structure of the protein kinase HIPK2 reveals a unique architecture of its CMGC-insert region.
Agnew, C., Liu, L., Liu, S., Xu, W., You, L., Yeung, W., Kannan, N., Jablons, D., Jura, N.(2019) J Biol Chem 294: 13545-13559
- PubMed: 31341017
- DOI: https://doi.org/10.1074/jbc.RA119.009725
- Primary Citation of Related Structures:
6P5S - PubMed Abstract:
The homeodomain-interacting protein kinase (HIPK) family is comprised of four nuclear protein kinases, HIPK1-4. HIPK proteins phosphorylate a diverse range of transcription factors involved in cell proliferation, differentiation, and apoptosis. HIPK2, thus far the best-characterized member of this largely understudied family of protein kinases, plays a role in the activation of p53 in response to DNA damage. Despite this tumor-suppressor function, HIPK2 is also found overexpressed in several cancers, and its hyperactivation causes chronic fibrosis. There are currently no structures of HIPK2 or of any other HIPK kinase. Here, we report the crystal structure of HIPK2's kinase domain bound to CX-4945, a casein kinase 2α (CK2α) inhibitor currently in clinical trials against several cancers. The structure, determined at 2.2 Å resolution, revealed that CX-4945 engages the HIPK2 active site in a hybrid binding mode between that seen in structures of CK2α and Pim1 kinases. The HIPK2 kinase domain crystallized in the active conformation, which was stabilized by phosphorylation of the activation loop. We noted that the overall kinase domain fold of HIPK2 closely resembles that of evolutionarily related dual-specificity tyrosine-regulated kinases (DYRKs). Most significant structural differences between HIPK2 and DYRKs included an absence of the regulatory N-terminal domain and a unique conformation of the CMGC-insert region and of a newly defined insert segment in the αC-β4 loop. This first crystal structure of HIPK2 paves the way for characterizing the understudied members of the HIPK family and for developing HIPK2-directed therapies for managing cancer and fibrosis.
Organizational Affiliation:
Cardiovascular Research Institute, University of California San Francisco, San Francisco, California 94158.