Comprehensive analysis of protein-protein interactions between MbtH-like protein FscK and adenylation domains in nonribosomal biosynthesis of Fuscachelins.
Bruner, S.D., Zagulyaeva, A.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MbtH-like protein | 81 | Thermobifida fusca YX | Mutation(s): 0 Gene Names: Tfu_1863 | 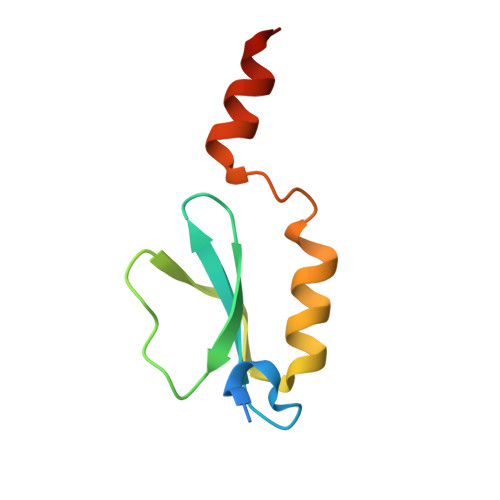 | |
UniProt | |||||
Find proteins for Q47NS3 (Thermobifida fusca (strain YX)) Explore Q47NS3 Go to UniProtKB: Q47NS3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q47NS3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| adenylation domain of Fuscachelin synthetase component H | 557 | Thermobifida fusca YX | Mutation(s): 0 Gene Names: Tfu_1866 | 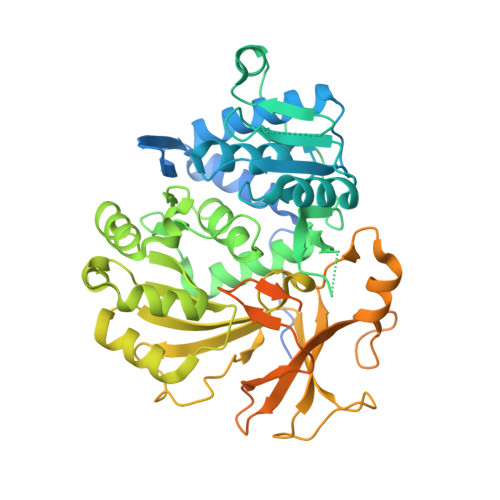 | |
UniProt | |||||
Find proteins for Q47NS0 (Thermobifida fusca (strain YX)) Explore Q47NS0 Go to UniProtKB: Q47NS0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q47NS0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SRP Query on SRP | C [auth B] | SERYL ADENYLATE C13 H19 N6 O9 P UVSYURUCZPPUQD-MACXSXHHSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 110.45 | α = 90 |
| b = 68.76 | β = 104.81 |
| c = 86.35 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| XDS | data reduction |
| XSCALE | data scaling |
| PHASER | phasing |