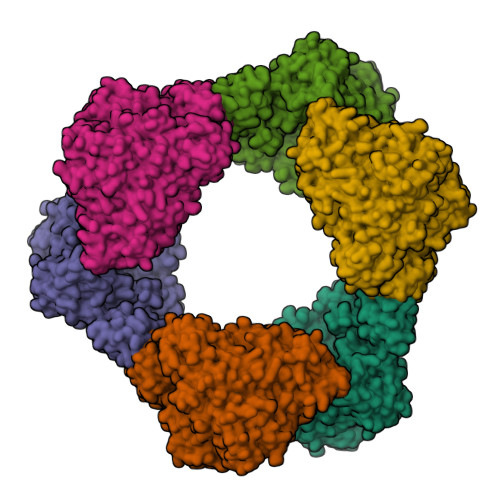
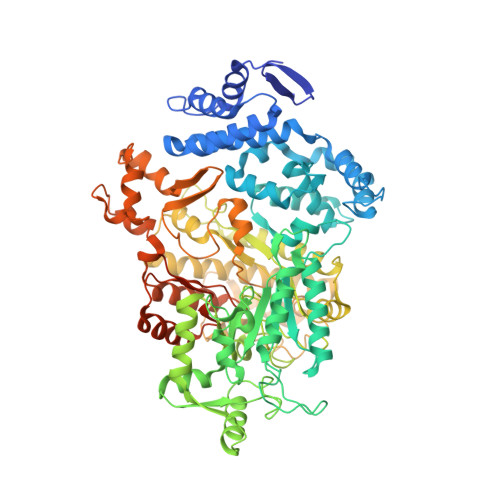
3.3- angstrom resolution cryo-EM structure of human ribonucleotide reductase with substrate and allosteric regulators bound.
Brignole, E.J., Tsai, K.L., Chittuluru, J., Li, H., Aye, Y., Penczek, P.A., Stubbe, J., Drennan, C.L., Asturias, F.(2018) Elife 7
- PubMed: 29460780
- DOI: https://doi.org/10.7554/eLife.31502
- Primary Citation of Related Structures:
6AUI - PubMed Abstract:
Ribonucleotide reductases (RNRs) convert ribonucleotides into deoxyribonucleotides, a reaction essential for DNA replication and repair. Human RNR requires two subunits for activity, the α subunit contains the active site, and the β subunit houses the radical cofactor. Here, we present a 3.3-Å resolution structure by cryo-electron microscopy (EM) of a dATP-inhibited state of human RNR. This structure, which was determined in the presence of substrate CDP and allosteric regulators ATP and dATP, has three α 2 units arranged in an α 6 ring. At near-atomic resolution, these data provide insight into the molecular basis for CDP recognition by allosteric specificity effectors dATP/ATP. Additionally, we present lower-resolution EM structures of human α 6 in the presence of both the anticancer drug clofarabine triphosphate and β 2 . Together, these structures support a model for RNR inhibition in which β 2 is excluded from binding in a radical transfer competent position when α exists as a stable hexamer.
Organizational Affiliation:
Howard Hughes Medical Institute, Massachusetts Institute of Technology, Cambridge, United States.