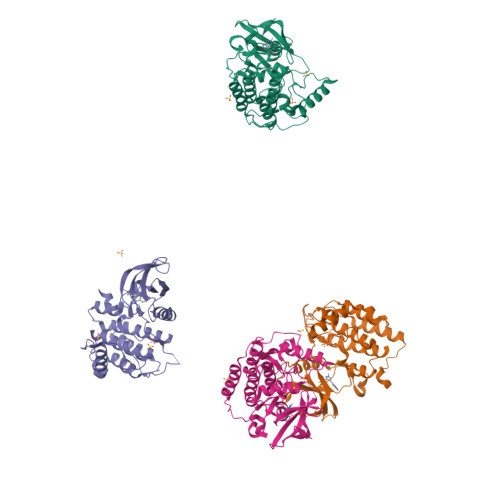
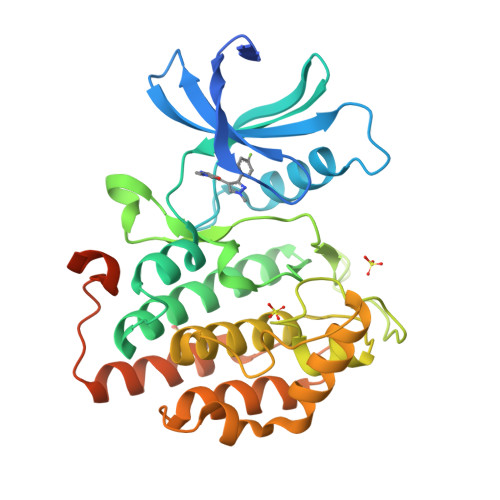
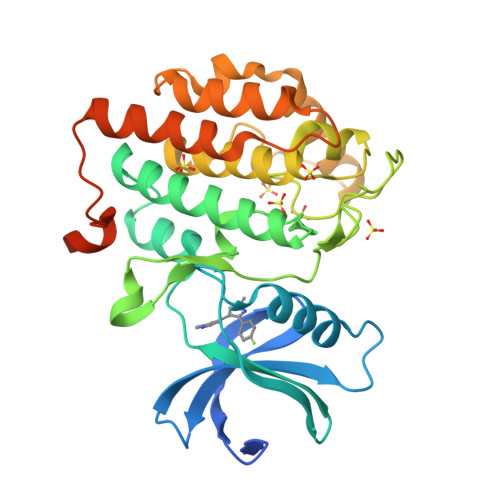
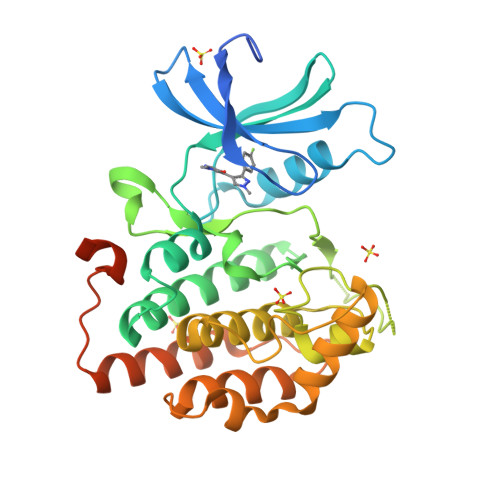
Identification and Profiling of a Selective and Brain Penetrant Radioligand for in Vivo Target Occupancy Measurement of Casein Kinase 1 (CK1) Inhibitors.
Wager, T.T., Galatsis, P., Chandrasekaran, R.Y., Butler, T.W., Li, J., Zhang, L., Mente, S., Subramanyam, C., Liu, S., Doran, A.C., Chang, C., Fisher, K., Grimwood, S., Hedde, J.R., Marconi, M., Schildknegt, K.(2017) ACS Chem Neurosci 8: 1995-2004
- PubMed: 28609096
- DOI: https://doi.org/10.1021/acschemneuro.7b00155
- Primary Citation of Related Structures:
5W4W - PubMed Abstract:
To enable the clinical development of our CNS casein kinase 1 delta/epsilon (CK1δ/ε) inhibitor project, we investigated the possibility of developing a CNS positron emission tomography (PET) radioligand. For this effort, we focused our design and synthesis efforts on the initial CK1δ/ε inhibitor HTS hits with the goal of identifying a compound that would fulfill a set of recommended PET ligand criteria. We identified [ 3 H]PF-5236216 (9) as a tool ligand that meets most of the key CNS PET attributes including high CNS MPO PET desirability score and kinase selectivity, CNS penetration, and low nonspecific binding. We further used [ 3 H]-9 to determine the binding affinity for PF-670462, a literature CK1δ/ε inhibitor tool compound. Lastly, [ 3 H]-9 was used to measure in vivo target occupancy (TO) of PF-670462 in mouse and correlated TO with CK1δ/ε in vivo pharmacology (circadian rhythm modulation).
Organizational Affiliation:
Worldwide Medicinal Chemistry, Pfizer Worldwide Research and Development , 1 Portland, Cambridge, Massachusetts 02139, United States.