Broad-Spectrum Kinase Profiling in Live Cells with Lysine-Targeted Sulfonyl Fluoride Probes.
Zhao, Q., Ouyang, X., Wan, X., Gajiwala, K.S., Kath, J.C., Jones, L.H., Burlingame, A.L., Taunton, J.(2017) J Am Chem Soc 139: 680-685
- PubMed: 28051857
- DOI: https://doi.org/10.1021/jacs.6b08536
- Primary Citation of Related Structures:
5K9I, 5U8L - PubMed Abstract:
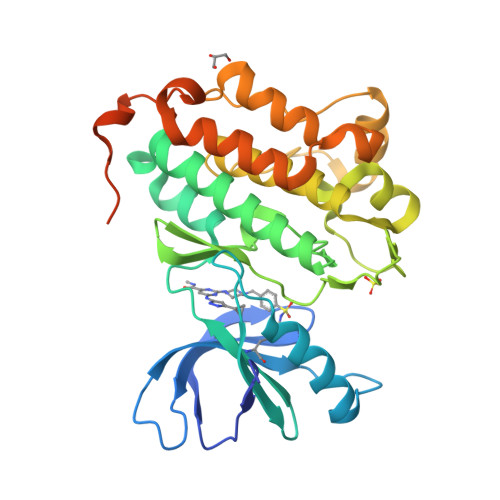
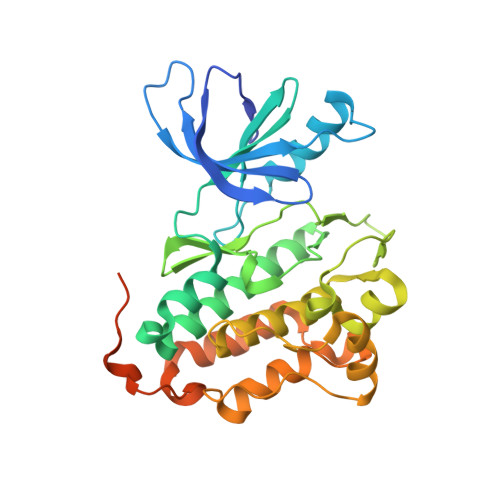
Protein kinases comprise a large family of structurally related enzymes. A major goal in kinase-inhibitor development is to selectively engage the desired kinase while avoiding myriad off-target kinases. However, quantifying inhibitor interactions with multiple endogenous kinases in live cells remains an unmet challenge. Here, we report the design of sulfonyl fluoride probes that covalently label a broad swath of the intracellular kinome with high efficiency. Protein crystallography and mass spectrometry confirmed a chemoselective reaction between the sulfonyl fluoride and a conserved lysine in the ATP binding site. Optimized probe 2 (XO44) covalently modified up to 133 endogenous kinases, efficiently competing with high intracellular concentrations of ATP. We employed probe 2 and label-free mass spectrometry to quantify intracellular kinase engagement by the approved drug, dasatinib. The data revealed saturable dasatinib binding to a small subset of kinase targets at clinically relevant concentrations, highlighting the utility of lysine-targeted sulfonyl fluoride probes in demanding chemoproteomic applications.
Organizational Affiliation:
Worldwide Research and Development, Pfizer , San Diego, California 92121, United States.