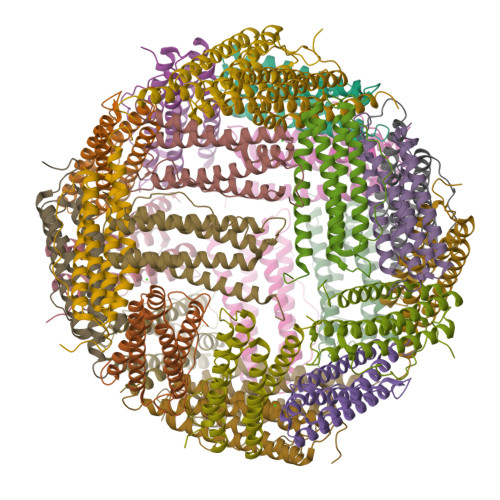
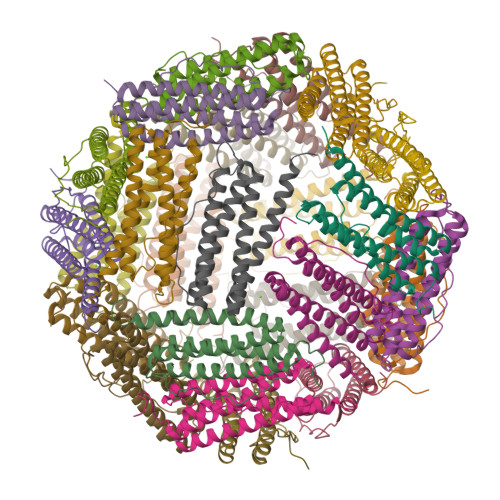
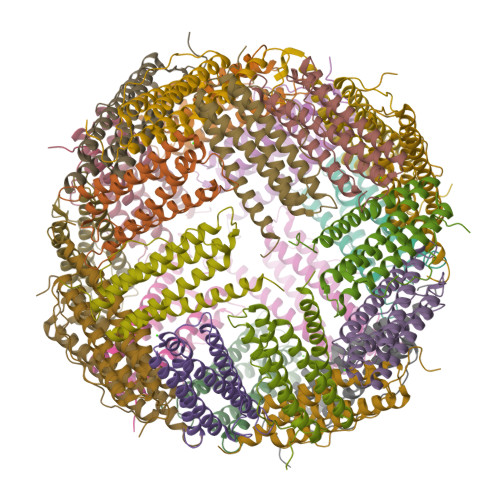
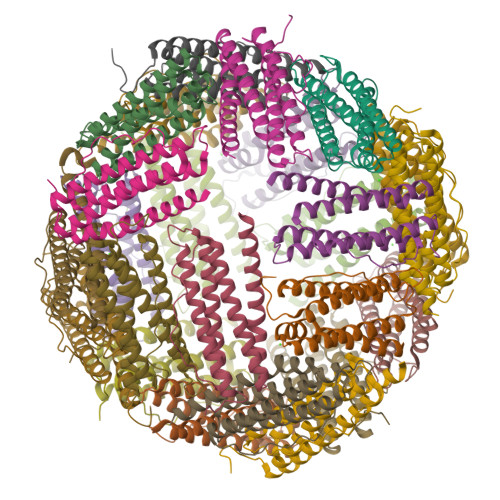
Unraveling of the E-helices and disruption of 4-fold pores are associated with iron mishandling in a mutant ferritin causing neurodegeneration
Baraibar, M.A., Muhoberac, B.B., Garringer, H.J., Hurley, T.D., Vidal, R.(2010) J Biological Chem 285: 1950-1956
- PubMed: 19923220
- DOI: https://doi.org/10.1074/jbc.M109.042986
- Primary Citation of Related Structures:
4V6B - PubMed Abstract:
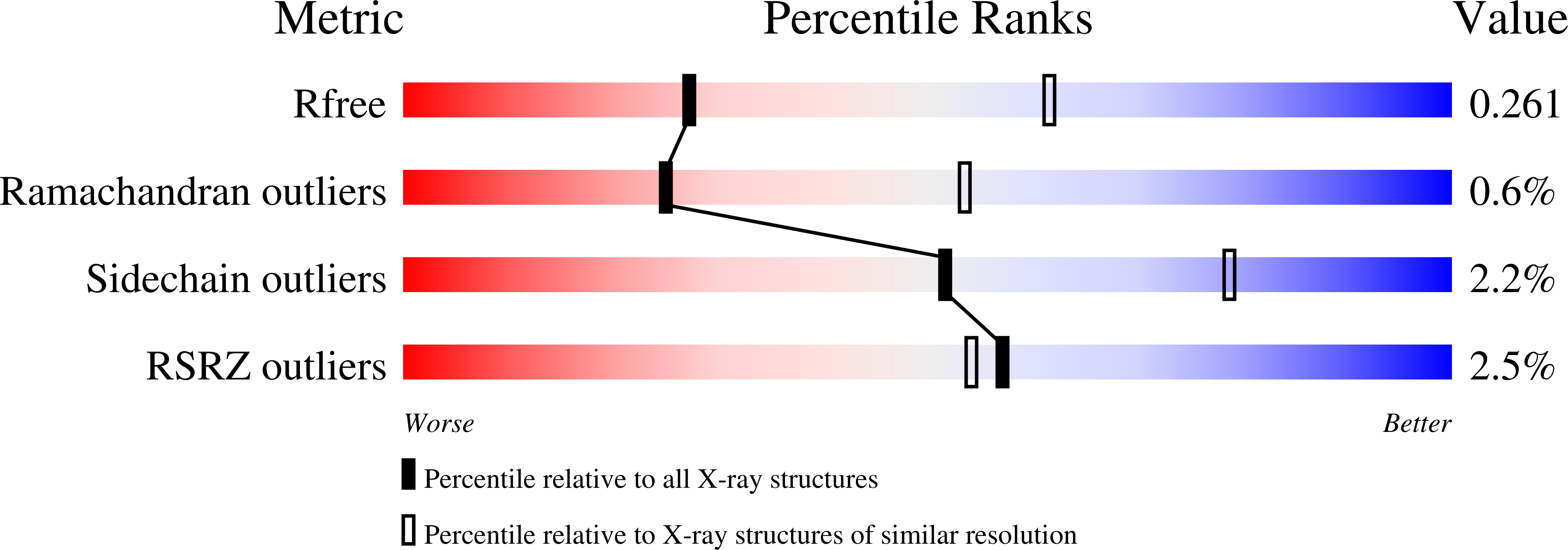
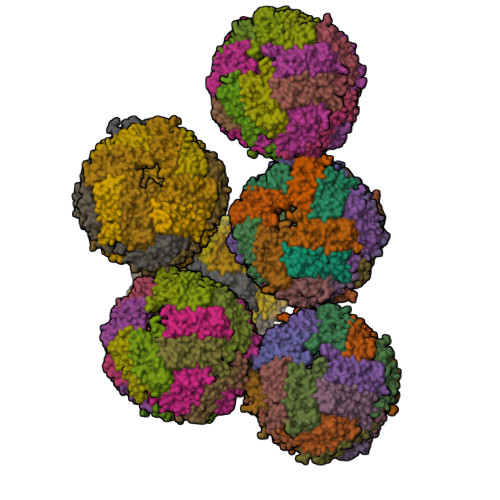
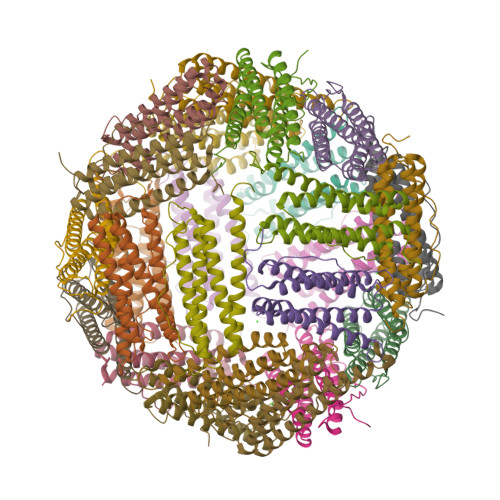
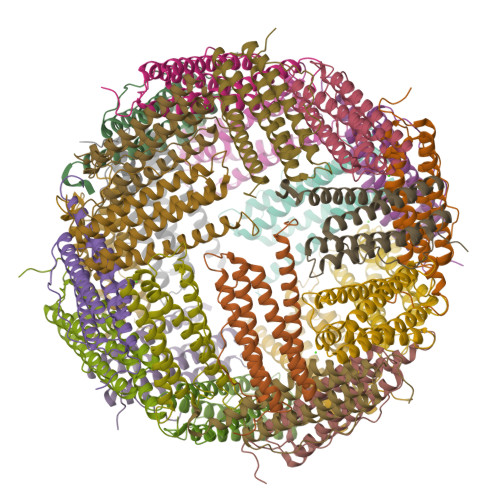
Mutations in the coding sequence of the ferritin light chain (FTL) gene cause a neurodegenerative disease known as neuroferritinopathy or hereditary ferritinopathy, which is characterized by the presence of intracellular inclusion bodies containing the mutant FTL polypeptide and by abnormal accumulation of iron in the brain. Here, we describe the x-ray crystallographic structure and report functional studies of ferritin homopolymers formed from the mutant FTL polypeptide p.Phe167SerfsX26, which has a C terminus that is altered in amino acid sequence and length. The structure was determined and refined to 2.85 A resolution and was very similar to the wild type between residues Ile-5 and Arg-154. However, instead of the E-helices normally present in wild type ferritin, the C-terminal sequences of all 24 mutant subunits showed substantial amounts of disorder, leading to multiple C-terminal polypeptide conformations and a large disruption of the normally tiny 4-fold axis pores. Functional studies underscored the importance of the mutant C-terminal sequence in iron-induced precipitation and revealed iron mishandling by soluble mutant FTL homopolymers in that only wild type incorporated iron when in direct competition in solution with mutant ferritin. Even without competition, the amount of iron incorporation over the first few minutes differed severalfold. Our data suggest that disruption at the 4-fold pores may lead to direct iron mishandling through attenuated iron incorporation by the soluble form of mutant ferritin and that the disordered C-terminal polypeptides may play a major role in iron-induced precipitation and formation of ferritin inclusion bodies in hereditary ferritinopathy.
Organizational Affiliation:
Department of Pathology and Laboratory Medicine, Indiana University School of Medicine, Indianapolis, Indiana 46202, USA.