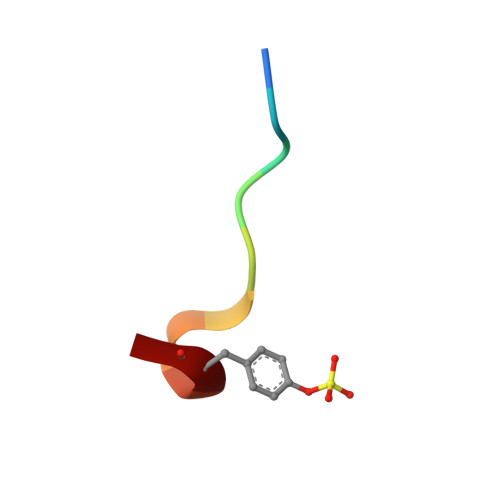
Identification of Structure-Kinetic and Structure-Thermodynamic Relationships for Thrombin Inhibitors.
Winquist, J., Geschwindner, S., Xue, Y., Gustavsson, L., Musil, D., Deinum, J., Danielson, U.H.(2013) Biochemistry 52: 613
- PubMed: 23290007
- DOI: https://doi.org/10.1021/bi301333z
- Primary Citation of Related Structures:
4BAH, 4BAK, 4BAM, 4BAN, 4BAO, 4BAQ - PubMed Abstract:
To improve our understanding of drug-target interactions, we explored the effect of introducing substituted amine residues with increased chain length in the P3 residue of the thrombin inhibitor melagatran. Inhibition, kinetic, and thermodynamic data obtained via stopped-flow spectroscopy (SF), isothermal microcalorimetry (ITC), and surface plasmon resonance (SPR) biosensor analysis were interpreted with the help of X-ray crystal structures of the enzyme-inhibitor complexes. The association rate became faster when the lipophilicity of the inhibitors was increased. This was coupled to an increased enthalpic component and a corresponding decreased entropic component. The dissociation rates were reduced with an increase in chain length, with only a smaller increase and a decrease in the enthalpic and entropic components, respectively. Overall, the affinity increased with an increase in chain length, with similar changes in the enthalpic and entropic components. ITC analysis confirmed the equilibrium data from SPR analysis, showing that the interaction of melagatran was the most enthalpy-driven interaction. Structural analysis of the thrombin-inhibitor complex showed that the orientation of the P1 and P2 parts of the molecules was very similar, but that there were significant differences in the interaction between the terminal part of the P3 side chain and the binding pocket. A combination of charge repulsion, H-bonds, and hydrophobic interactions could be used to explain the observed kinetic and thermodynamic profiles for the ligands. In conclusion, changes in the structure of a lead compound can have significant effects on its interaction with the target that translate directly into kinetic and thermodynamic effects. In contrast to what may be intuitively expected, hydrogen bond formation and breakage are not necessarily reflected in enthalpy gains and losses, respectively.
Organizational Affiliation:
Department of Chemistry-BMC, Uppsala University, SE-751 23 Uppsala, Sweden.