N-aryl-benzimidazolones as novel small molecule HSP90 inhibitors.
Bruncko, M., Tahir, S.K., Song, X., Chen, J., Ding, H., Huth, J.R., Jin, S., Judge, R.A., Madar, D.J., Park, C.H., Park, C.M., Petros, A.M., Tse, C., Rosenberg, S.H., Elmore, S.W.(2010) Bioorg Med Chem Lett 20: 7503-7506
- PubMed: 21106457
- DOI: https://doi.org/10.1016/j.bmcl.2010.10.010
- Primary Citation of Related Structures:
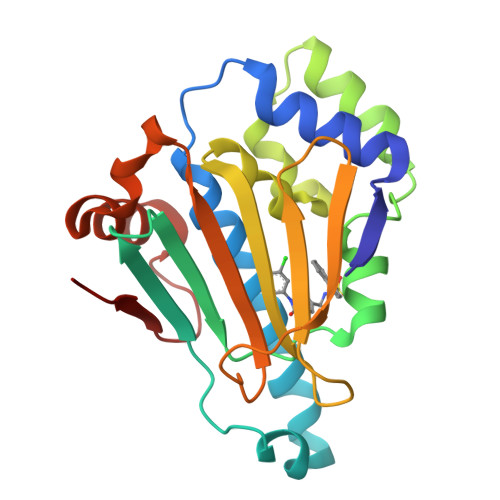
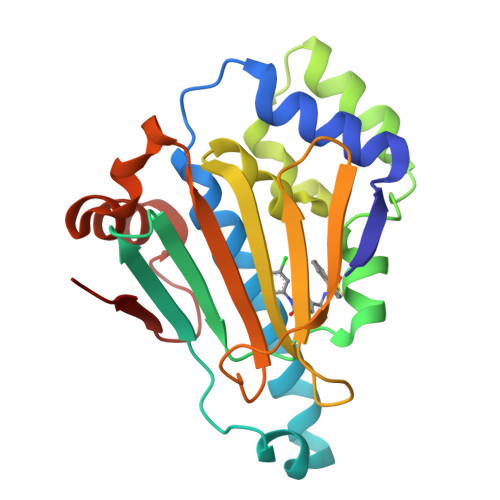
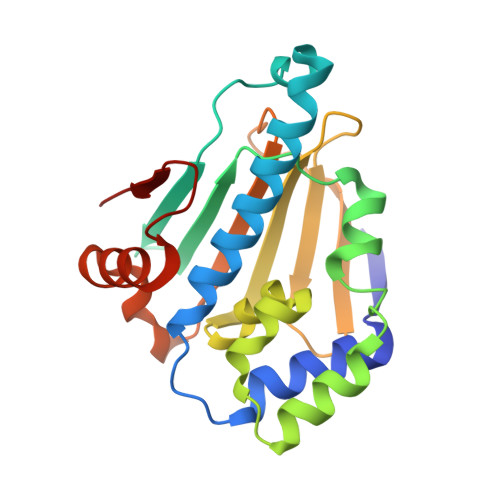
3OW6, 3OWB, 3OWD - PubMed Abstract:
We describe the development of a novel series of N-aryl-benzimidazolone HSP90 inhibitors (9) targeting the N-terminal ATP-ase site. SAR development was influenced by structure-based design based around X-ray structures of ligand bound HSP90 complexes. Lead compounds exhibited high binding affinities, ATP-ase inhibition and cellular client protein degradation.
Organizational Affiliation:
Cancer Research, Global Pharmaceutical R&D, Abbott Laboratories, Abbott Park, IL 60064-6101, USA. milan.bruncko@abbott.com