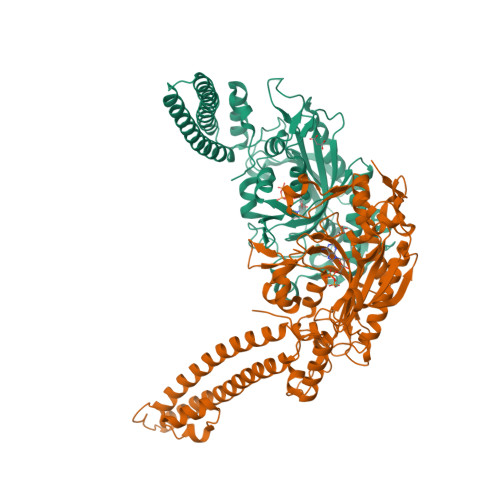
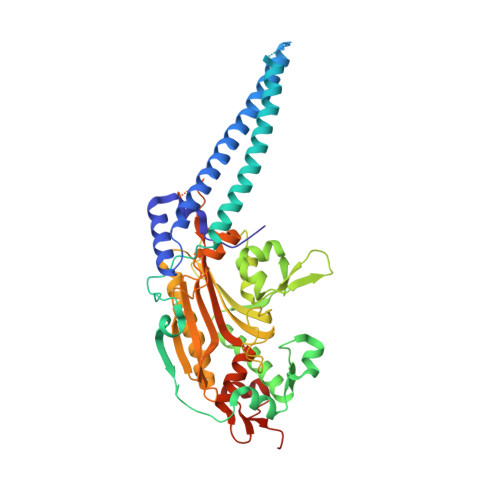
X-ray crystal structure of Seryl-tRNA synthetase from the eukaryotic parasite Trypanosoma brucei.
Larson, E.T., Zhang, L., Napuli, A., Mueller, N., Verlinde, C.L.M.J., Van Voorhis, W.C., Buckner, F.S., Fan, E., Hol, W.G.J., Merritt, E.A.To be published.