Crystal Structures of the Organomercurial Lyase MerB in Its Free and Mercury-bound Forms: INSIGHTS INTO THE MECHANISM OF METHYLMERCURY DEGRADATION
Lafrance-Vanasse, J., Lefebvre, M., Di Lello, P., Sygusch, J., Omichinski, J.G.(2009) J Biological Chem 284: 938-944
- PubMed: 19004822
- DOI: https://doi.org/10.1074/jbc.M807143200
- Primary Citation of Related Structures:
3F0O, 3F0P, 3F2F, 3F2G, 3F2H - PubMed Abstract:
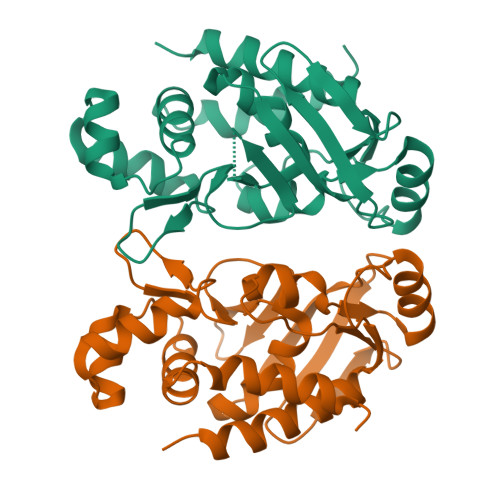
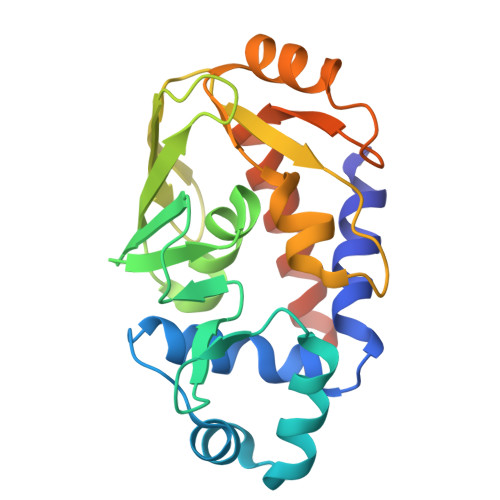
Bacteria resistant to methylmercury utilize two enzymes (MerA and MerB) to degrade methylmercury to the less toxic elemental mercury. The crucial step is the cleavage of the carbon-mercury bond of methylmercury by the organomercurial lyase (MerB). In this study, we determined high resolution crystal structures of MerB in both the free (1.76-A resolution) and mercury-bound (1.64-A resolution) states. The crystal structure of free MerB is very similar to the NMR structure, but important differences are observed when comparing the two structures. In the crystal structure, an amino-terminal alpha-helix that is not present in the NMR structure makes contact with the core region adjacent to the catalytic site. This interaction between the amino-terminal helix and the core serves to bury the active site of MerB. The crystal structures also provide detailed insights into the mechanism of carbon-mercury bond cleavage by MerB. The structures demonstrate that two conserved cysteines (Cys-96 and Cys-159) play a role in substrate binding, carbon-mercury bond cleavage, and controlled product (ionic mercury) release. In addition, the structures establish that an aspartic acid (Asp-99) in the active site plays a crucial role in the proton transfer step required for the cleavage of the carbon-mercury bond. These findings are an important step in understanding the mechanism of carbon-mercury bond cleavage by MerB.
Organizational Affiliation:
Department of Biochemistry, Université de Montréal, Montréal, Québec H3C 3J7, Canada.