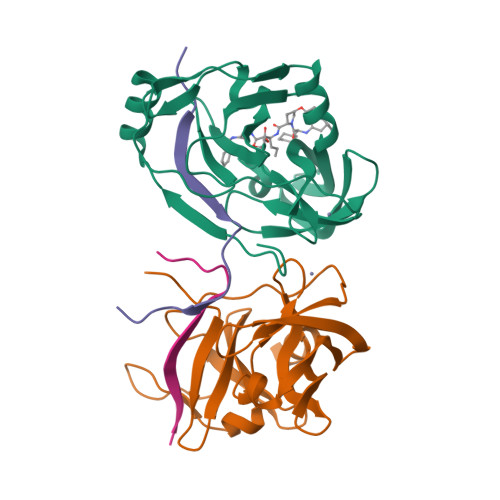
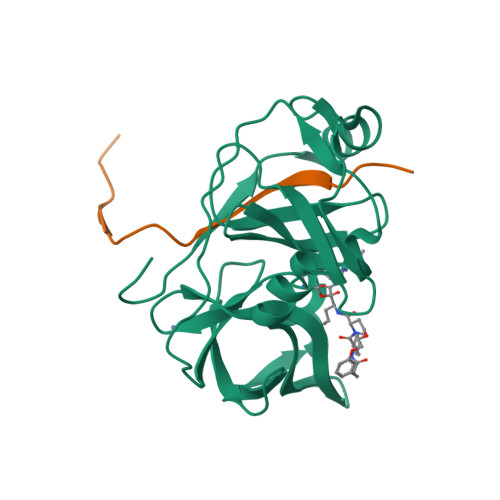
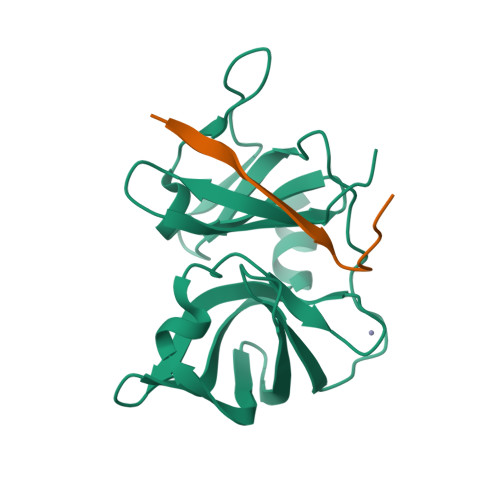
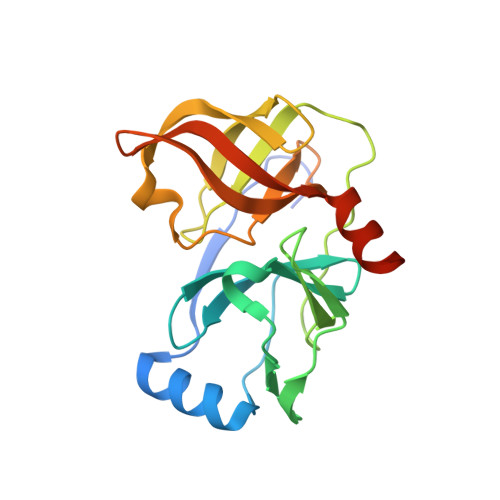
P2-P4 macrocyclic inhibitors of hepatitis C virus NS3-4A serine protease.
Arasappan, A., Njoroge, F.G., Chen, K.X., Venkatraman, S., Parekh, T.N., Gu, H., Pichardo, J., Butkiewicz, N., Prongay, A., Madison, V., Girijavallabhan, V.(2006) Bioorg Med Chem Lett 16: 3960-3965
- PubMed: 16730985
- DOI: https://doi.org/10.1016/j.bmcl.2006.05.022
- Primary Citation of Related Structures:
2GVF - PubMed Abstract:
Synthesis and HCV NS3 serine protease inhibitory activity of 4-hydroxyproline derived macrocyclic inhibitors and SAR around this macrocyclic core is described in this communication. X-ray structure of inhibitor 38 bound to the protease is discussed.
Organizational Affiliation:
Schering Plough Research Institute, 2015 Galloping Hill Road, Kenilworth, NJ 07033, USA. ashok.arasappan@spcorp.com