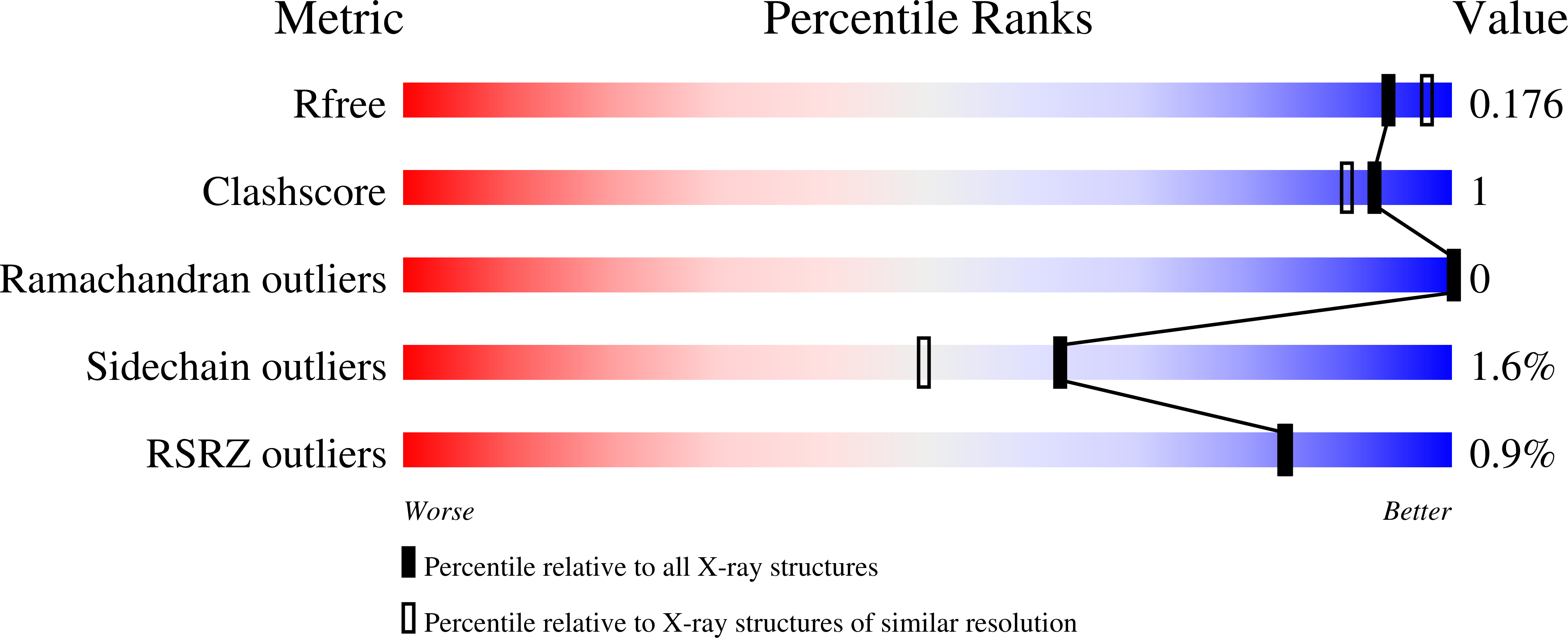
X-ray and NMR characterization of covalent complexes of trypsin, borate, and alcohols.
Transue, T.R., Krahn, J.M., Gabel, S.A., DeRose, E.F., London, R.E.(2004) Biochemistry 43: 2829-2839
- PubMed: 15005618
- DOI: https://doi.org/10.1021/bi035782y
- Primary Citation of Related Structures:
1S5S, 1S6F, 1S6H, 1S81, 1S82, 1S83, 1S84, 1S85 - PubMed Abstract:
An understanding of the physiological and toxicological properties of borate and the utilization of boronic acids in drug development require a basic understanding of borate-enzyme chemistry. We report here the extension of our recent NMR studies indicating the formation of a ternary borate-alcohol-trypsin complex. Crystallographic and solution state NMR studies of porcine trypsin were performed in the presence of borate and either of three alcohols designed to bind to the S1 affinity subsite: 4-aminobutanol, guanidine-3-propanol, and 4-hydroxymethylbenzamidine. Quaternary complexes of trypsin, borate, S1-binding alcohol, and ethylene glycol (a cryoprotectant), as well as a ternary trypsin, borate, and ethylene glycol complex have been observed in the crystalline state. Borate forms ester bonds to Ser195, ethylene glycol (two bonds), and the S1-binding alcohol (if present). Spectra from (1)H and (11)B NMR studies confirm that these complexes also exist in solution and also provide evidence for the formation of ternary trypsin, borate, and S1-subsite alcohol complexes which are not observed in the crystals using our experimental protocols. Analysis of eight crystal structures indicates that formation of an active site borate complex is in all cases accompanied by a significant (approximately 4%) increase in the b-axis dimension of the unit cell. Presumably, our inability to observe the ternary complexes in the crystalline state arises from the lower stability of these complexes and consequent inability to overcome the constraints imposed by the lattice contacts. A mechanism for the coupling of the lattice contacts with the active site that involves a conformational rearrangement of Gln192 is suggested. The structures presented here represent the first crystallographic demonstration of covalent binding of an enzyme by borate.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, 111 T.W. Alexander Drive, Research Triangle Park, North Carolina 27709, USA.