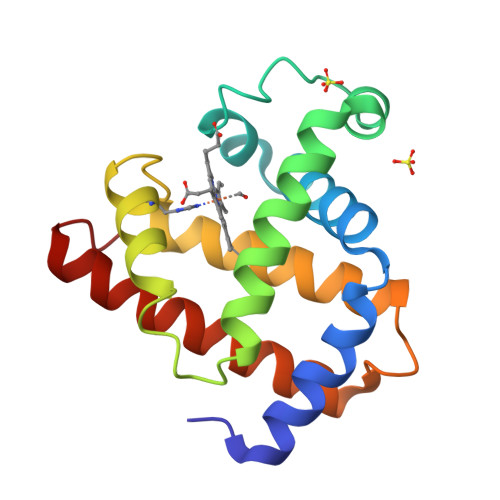
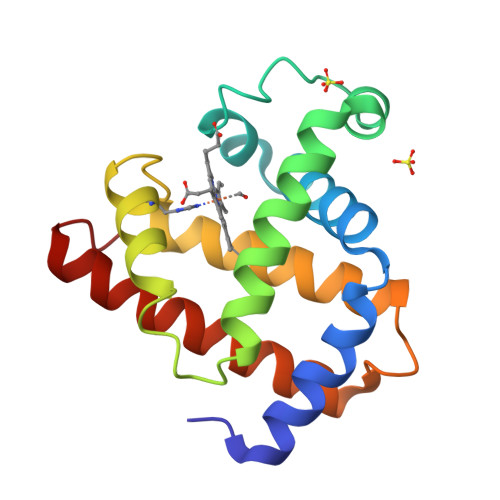
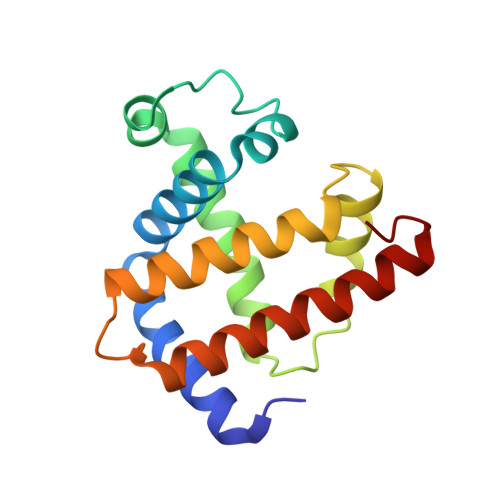
Photolysis-induced structural changes in single crystals of carbonmonoxy myoglobin at 40 K.
Teng, T.Y., Srajer, V., Moffat, K.(1994) Nat Struct Biol 1: 701-705
- PubMed: 7634074
- DOI: https://doi.org/10.1038/nsb1094-701
- Primary Citation of Related Structures:
1AJG, 1AJH - PubMed Abstract:
Myoglobin's reversible binding of oxygen is a model for studies of protein control of ligand binding and discrimination. Protein relaxation and geminate ligand rebinding subsequent to ligand photodissociation have been studied extensively by a variety of techniques. The ps to ns time scales for these processes are still much shorter than the ms time resolution of X-ray diffraction experiments, but it may be possible to trap these intermediates at low temperatures. We report here an X-ray diffraction investigation of structural changes induced by photolysis of carbonmonoxy myoglobin crystals at 40 K. Our results provide a structural basis for the interpretation of ambient and low temperature spectroscopic observations and molecular dynamics simulations of the ligand photodissociation and binding processes in haem proteins.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, University of Chicago, IL 60637, USA.