Full Text |
QUERY: Structure Author = "Lee, K.K." | MyPDB Login | Search API |
| Search Summary | This query matches 9 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 9 of 9 Structures Page 1 of 1 Sort by
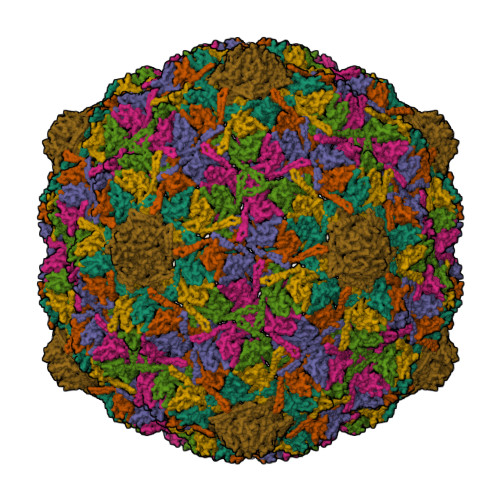
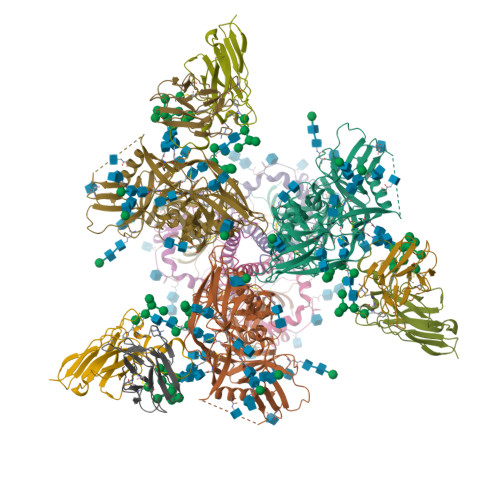
HK97 bacteriophage capsid Expansion Intermediate-II modelLee, K.K., Gan, L., Conway, J.F., Hendrix, R.W., Steven, A.C., Johnson, J.E. (2008) Structure 16: 1491-1502
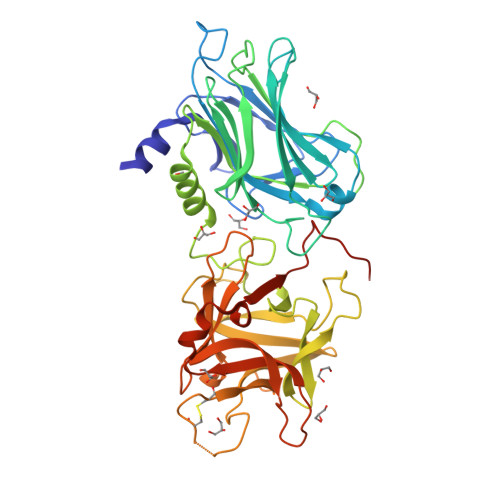
Crystal structure of Botulinum neurotoxin serotype D binding domain(2010) Biochem J 431: 207-216
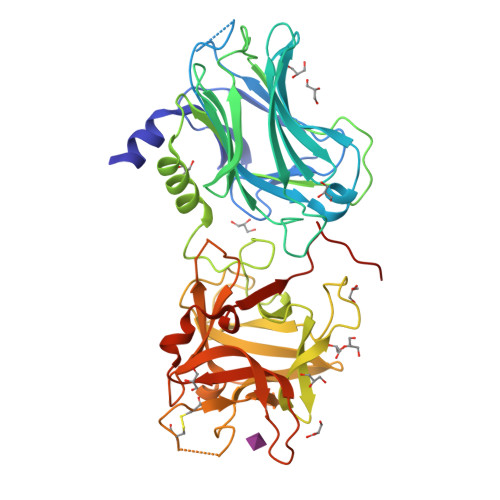
Crystal structure of Botulinum neurotoxin serotype D ligand binding domain in complex with N-Acetylneuraminic acid(2010) Biochem J 431: 207-216
HK97 Prohead I encapsidating inactive virally encoded proteaseHuang, R.K., Khayat, R., Lee, K.K., Gertsman, I., Duda, R.L., Hendrix, R.W., Johnson, J.E. (2011) J Mol Biology 408: 541-554
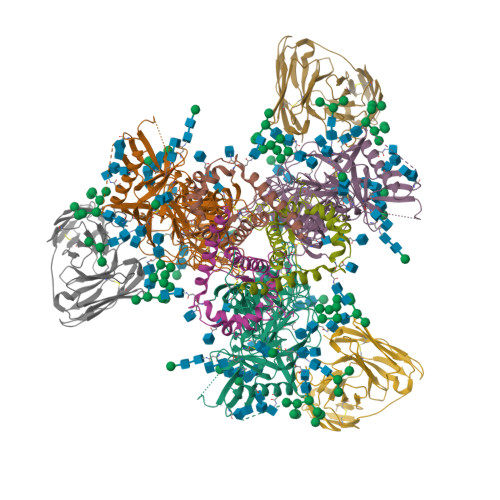
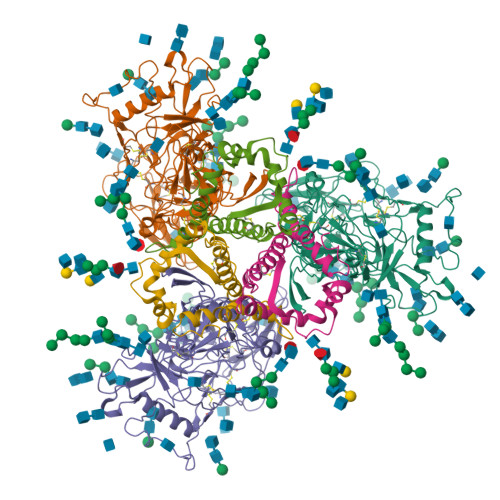
Cryo-EM structure of BG505.SOSIP.664 in complex with BF520.1 antigen binding fragmentWilliams, J.A., Lee, K.K., Overbaugh, J. (2019) Nat Commun 10: 2190-2190
Complex structure of HIV superinfection Fab QA013.2 and BG505.SOSIP.664Mangala Prasad, V., Shipley, M.M., Overbaugh, J.M., Lee, K.K. (2021) Elife 10:
Sub-tomogram averaged structure of HIV-1 Envelope protein in native membrane(2022) Cell 185: 641-653.e17
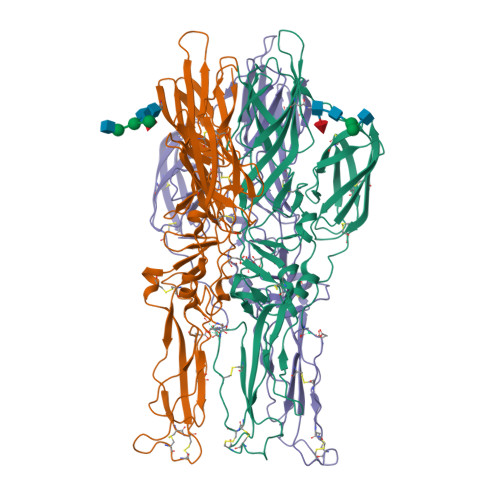
Fitted crystal structure of the homotrimer of fusion glycoprotein E1 from SFV into subtomogram averaged CHIKV E1 glycoprotein density(2022) Nat Commun 13: 4772-4772
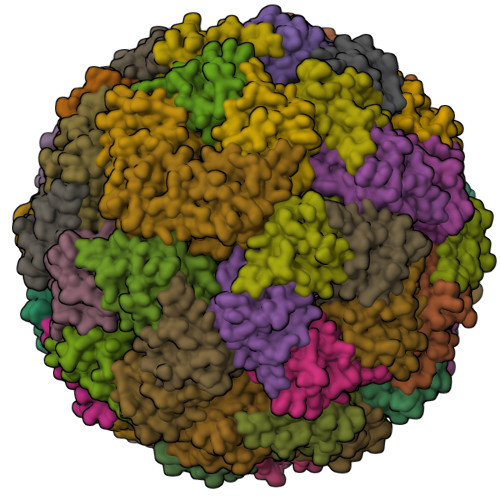
Cryo-EM structure of Lumazine synthase nanoparticle linked to VP8* antigen(2023) NPJ Vaccines 8: 190-190
1 to 9 of 9 Structures Page 1 of 1 Sort by |