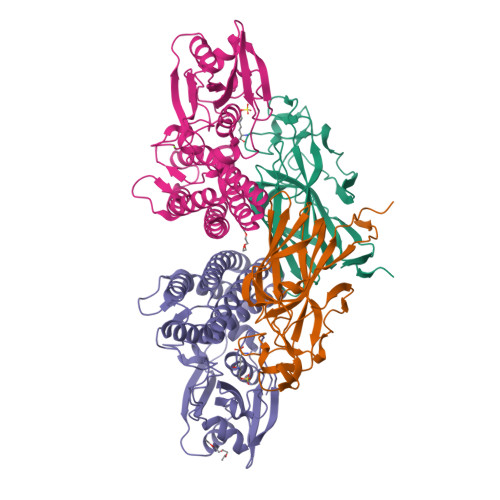
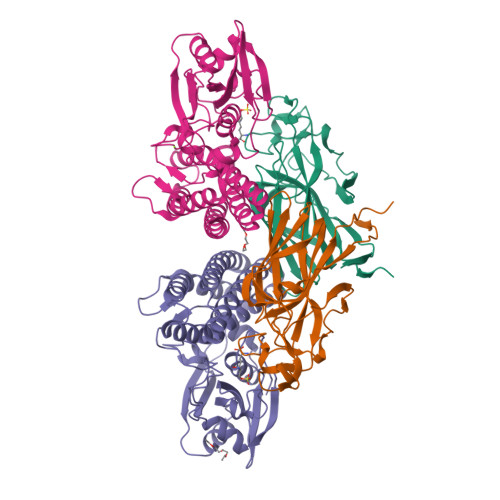
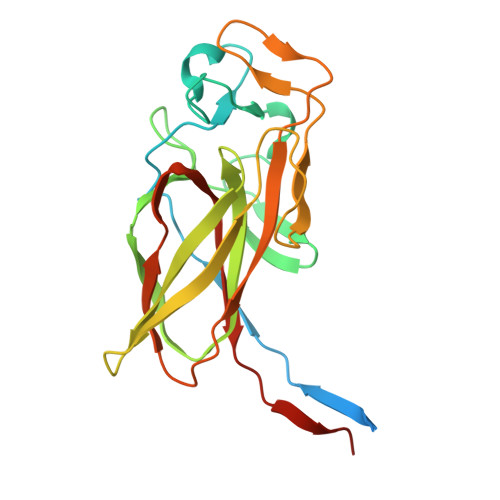
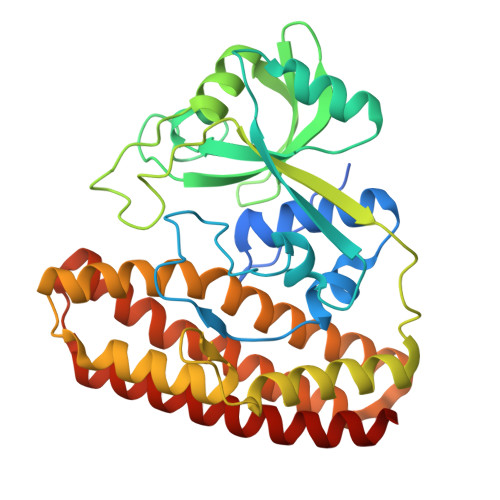
Molecular Basis for Enzymatic Aziridine Formation via Sulfate Elimination.
Kurosawa, S., Hasebe, F., Okamura, H., Yoshida, A., Matsuda, K., Sone, Y., Tomita, T., Shinada, T., Takikawa, H., Kuzuyama, T., Kosono, S., Nishiyama, M.(2022) J Am Chem Soc 144: 16164-16170
- PubMed: 35998388
- DOI: https://doi.org/10.1021/jacs.2c07243
- Primary Citation of Related Structures:
7WUW, 7WUX - PubMed Abstract:
Natural products containing an aziridine ring, such as mitomycin C and azinomycin B, exhibit antitumor activities by alkylating DNA via their aziridine rings; however, the biosynthetic mechanisms underlying the formation of these rings have not yet been elucidated. We herein investigated the biosynthesis of vazabitide A, the structure of which is similar to that of azinomycin B, and demonstrated that Vzb10/11, with no similarities to known enzymes, catalyzed the formation of the aziridine ring via sulfate elimination. To elucidate the detailed reaction mechanism, crystallization of Vzb10/11 and the homologous enzyme, AziU3/U2, in the biosynthesis of azinomycin B was attempted, and the structure of AziU3/U2, which had a new protein fold overall, was successfully determined. The structural analysis revealed that these enzymes adjusted the dihedral angle between the amino group and the adjacent sulfate group of the substrate to almost 180° and enhanced the nucleophilicity of the C6-amino group temporarily, facilitating the S N 2-like reaction to form the aziridine ring. The present study reports for the first time the molecular basis for aziridine ring formation.
Organizational Affiliation:
Graduate School of Agricultural and Life Sciences, The University of Tokyo, 1-1-1, Yayoi, Bunkyo-ku, Tokyo 113-8657, Japan.