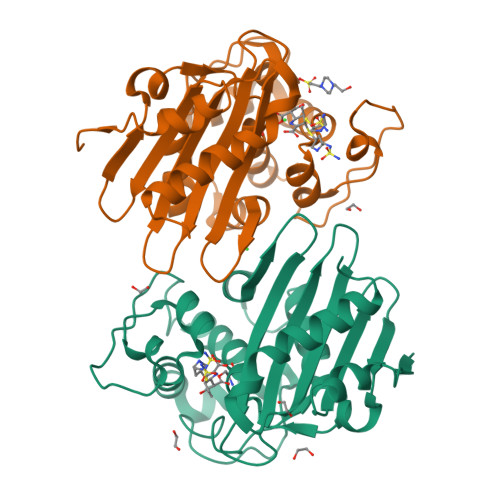
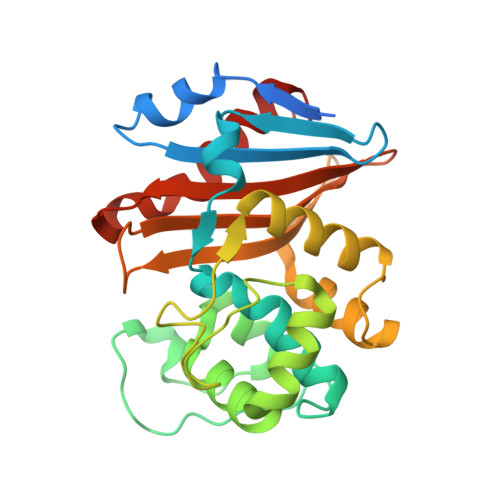
Structural Analysis of The OXA-48 Carbapenemase Bound to A "Poor" Carbapenem Substrate, Doripenem.
Papp-Wallace, K.M., Kumar, V., Zeiser, E.T., Becka, S.A., van den Akker, F.(2019) Antibiotics (Basel) 8
- PubMed: 31514291
- DOI: https://doi.org/10.3390/antibiotics8030145
- Primary Citation of Related Structures:
6PXX - PubMed Abstract:
Carbapenem-resistant Enterobacteriaceae are a significant threat to public health, and a major resistance determinant that promotes this phenotype is the production of the OXA-48 carbapenemase. The activity of OXA-48 towards carbapenems is a puzzling phenotype as its hydrolytic activity against doripenem is non-detectable. To probe the mechanistic basis for this observation, we determined the 1.5 Å resolution crystal structure of the deacylation deficient K73A variant of OXA-48 in complex with doripenem. Doripenem is observed in the Δ 1 R and Δ 1 S tautomeric states covalently attached to the catalytic S70 residue. Likely due to positioning of residue Y211, the carboxylate moiety of doripenem is making fewer hydrogen bonding/salt-bridge interactions with R250 compared to previously determined carbapenem OXA structures. Moreover, the hydroxyethyl side chain of doripenem is making van der Waals interactions with a key V120 residue, which likely affects the deacylation rate of doripenem. We hypothesize that positions V120 and Y211 play important roles in the carbapenemase profile of OXA-48. Herein, we provide insights for the further development of the carbapenem class of antibiotics that could render them less effective to hydrolysis by or even inhibit OXA carbapenemases.
Organizational Affiliation:
Department of Biochemistry, Case Western Reserve University, Cleveland, OH 44106, USA. krisztina.papp@va.gov.