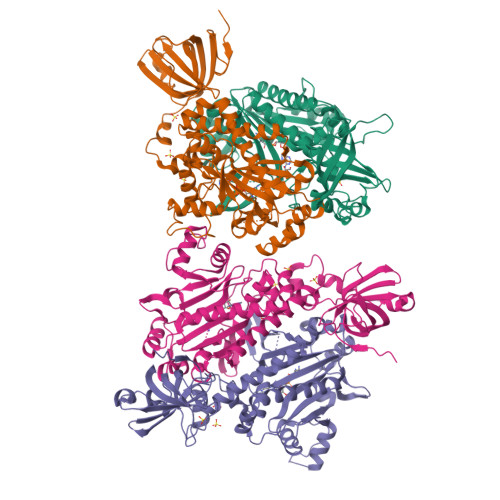
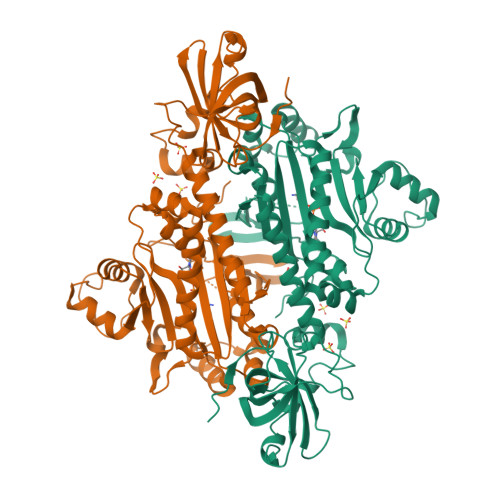
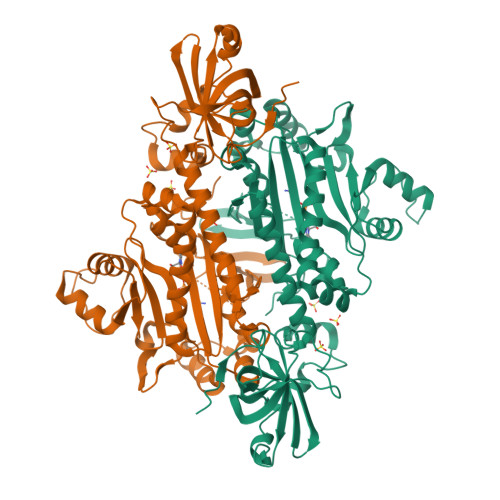
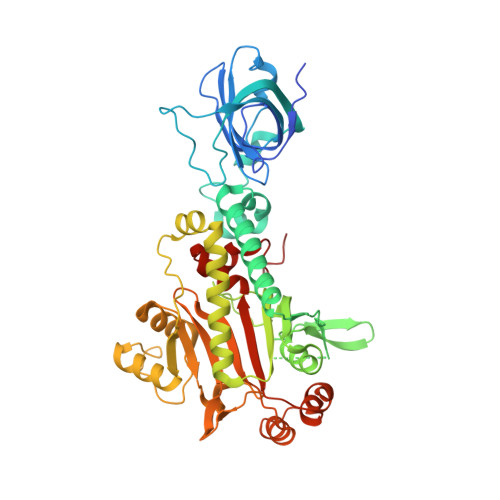
X-ray crystal structure of asparaginyl-tRNA synthetase from the eukaryotic human pathogen Entamoeba histolytica.
Larson, E.T., Kim, J.E., Napuli, A., Kelley, A., Castaneda, L., Verlinde, C.L.M.J., Van Voorhis, W.C., Buckner, F.S., Fan, E., Hol, W.G.J., Merritt, E.A.To be published.