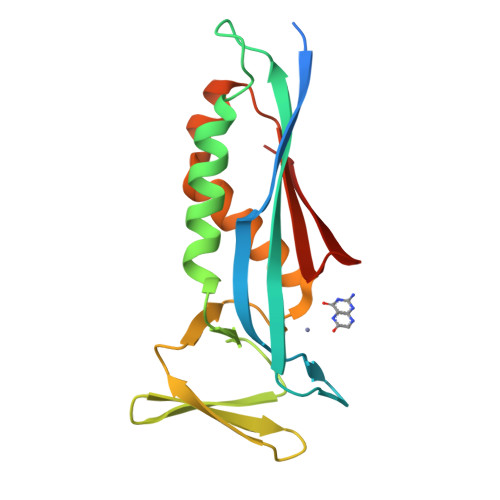
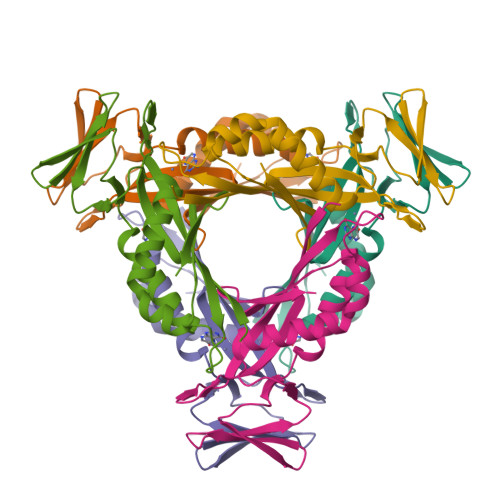
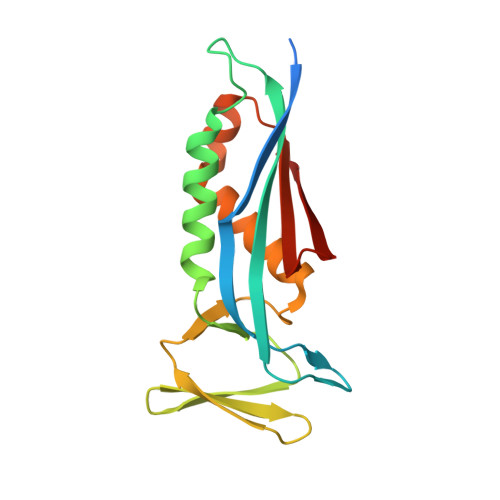
Structural analysis of the dual-functional 6-pyruvoyltetrahydropterin synthase from Malaria parasites.
Larson, E.T., Bosch, J., Kim, J.E., Mueller, N., Verlinde, C.L.M.J., Van Voorhis, W.C., Buckner, F.S., Fan, E., Hol, W.G.J., Merritt, E.A.To be published.