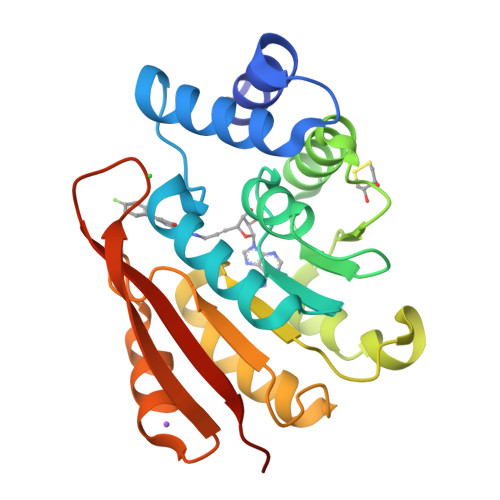
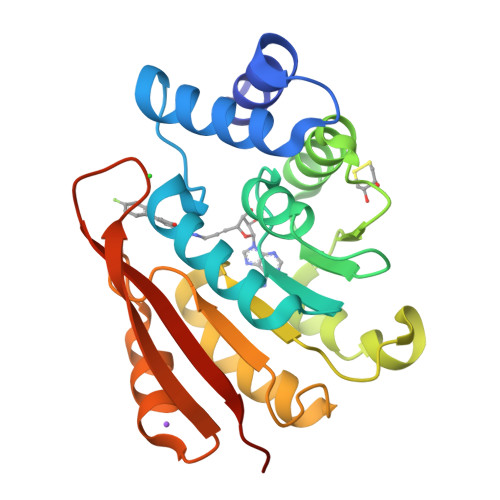
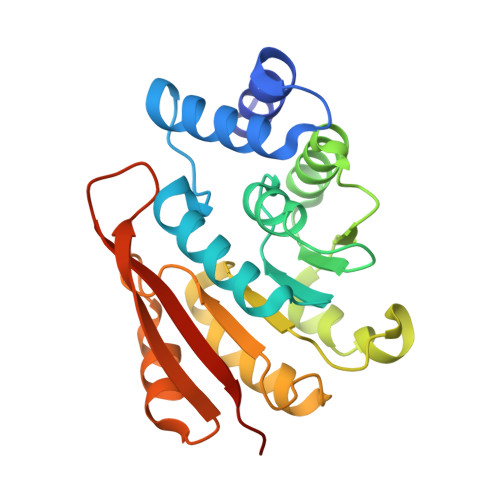
Molecular recognition at the active site of catechol-o-methyltransferase: energetically favorable replacement of a water molecule imported by a bisubstrate inhibitor.
Ellermann, M., Jakob-Roetne, R., Lerner, C., Borroni, E., Schlatter, D., Roth, D., Ehler, A., Rudolph, M.G., Diederich, F.(2009) Angew Chem Int Ed Engl 48: 9092-9096