Carboxy-Terminus Recruitment Induced by Substrate Binding in Eukaryotic Fructose Bis-phosphate Aldolases
Lafrance-Vanasse, J., Sygusch, J.(2007) Biochemistry 46: 9533-9540
- PubMed: 17661446
- DOI: https://doi.org/10.1021/bi700615r
- Primary Citation of Related Structures:
2QAP, 2QDG, 2QDH - PubMed Abstract:
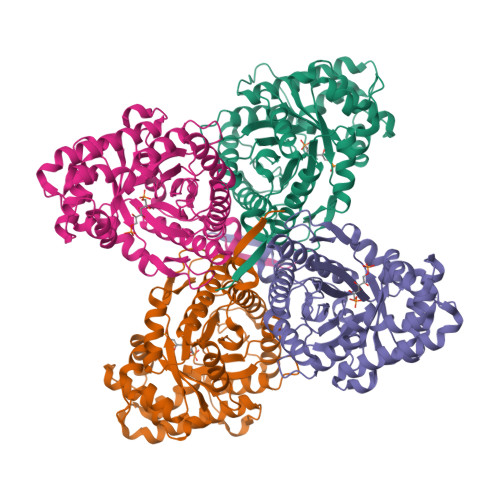
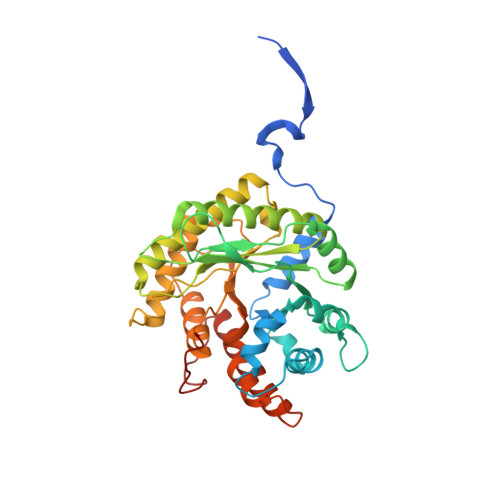
The crystal structures of Leishmania mexicana fructose-1,6-bis(phosphate) aldolase in complex with substrate and competitive inhibitor, mannitol-1,6-bis(phosphate), were solved to 2.2 A resolution. Crystallographic analysis revealed a Schiff base intermediate trapped in the native structure complexed with substrate while the inhibitor was trapped in a conformation mimicking the carbinolamine intermediate. Binding modes corroborated previous structures reported for rabbit muscle aldolase. Amino acid substitution of Gly-312 to Ala, adjacent to the P1-phosphate binding site and unique to trypanosomatids, did not perturb ligand binding in the active site. Ligand attachment ordered amino acid residues 359-367 of the C-terminal region (353-373) that was disordered beyond Asp-358 in the unbound structure, revealing a novel recruitment mechanism of this region by aldolases. C-Terminal peptide ordering is triggered by P1-phosphate binding that induces conformational changes whereby C-terminal Leu-364 contacts P1-phosphate binding residue Arg-313. C-Terminal region capture synergizes additional interactions with subunit surface residues, not perturbed by P1-phosphate binding, and stabilizes C-terminal attachment. Amino acid residues that participate in the capturing interaction are conserved among class I aldolases, indicating a general recruitment mechanism whereby C-terminal capture facilitates active site interactions in subsequent catalytic steps. Recruitment accelerates the enzymatic reaction by using binding energy to reduce configurational entropy during catalysis thereby localizing the conserved C-terminus tyrosine, which mediates proton transfer, proximal to the active site enamine.
Organizational Affiliation:
Département de Biochimie, Université de Montréal, Montréal, Québec H3C 3J7, Canada.