Development of radiofluorinated MLN-4760 derivatives for PET imaging of the SARS-CoV-2 entry receptor ACE2.
Wang, J., Beyer, D., Vaccarin, C., He, Y., Tanriver, M., Benoit, R., Deupi, X., Mu, L., Bode, J.W., Schibli, R., Muller, C.(2024) Eur J Nucl Med Mol Imaging 52: 9-21
- PubMed: 39066808
- DOI: https://doi.org/10.1007/s00259-024-06831-6
- Primary Citation of Related Structures:
9FMM - PubMed Abstract:
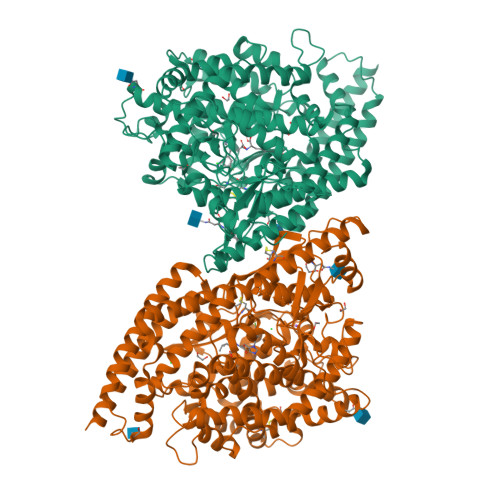
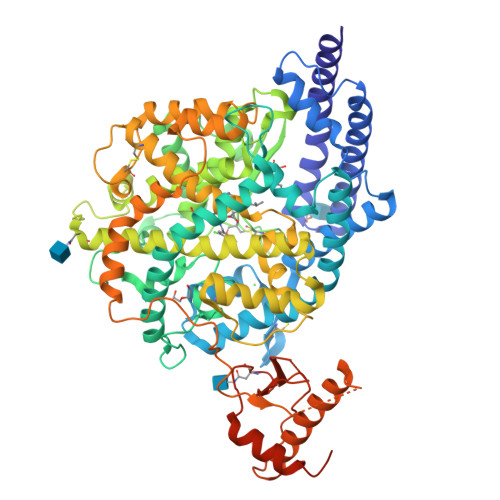
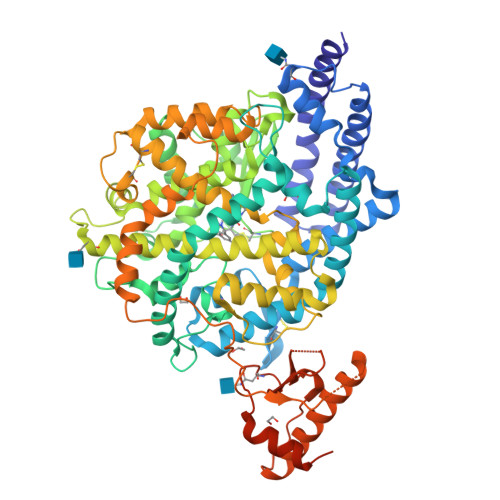
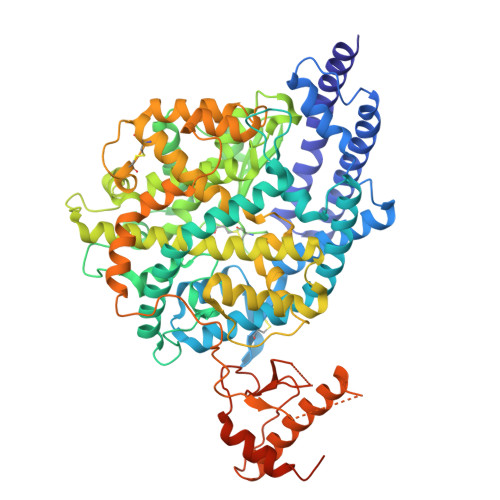
The angiotensin converting enzyme 2 (ACE2) plays a regulatory role in the cardiovascular system and serves SARS-CoV-2 as an entry receptor. The aim of this study was to synthesize and evaluate radiofluorinated derivatives of the ACE2 inhibitor MLN-4760. [ 18 F]F-MLN-4760 and [ 18 F]F-Aza-MLN-4760 were demonstrated to be suitable for non-invasive imaging of ACE2, potentially enabling a better understanding of its expression dynamics. Computational molecular modeling, based on the structures of human ACE2 (hACE2) and mouse ACE2 (mACE2), revealed that the ACE2-binding modes of F-MLN-4760 and F-Aza-MLN-4760 were similar to that of MLN-4760. Co-crystallization of the hACE2/F-MLN-4760 protein complex was performed for confirmation. Displacement experiments using [ 3 H]MLN-4760 enabled the determination of the binding affinities of the synthesized F-MLN-4760 and F-Aza-MLN-4760 to hACE2 expressed in HEK-ACE2 cells. Aryl trimethylstannane-based and pyridine-based radiofluorination precursors were synthesized and used for the preparation of the respective radiotracers. [ 18 F]F-MLN-4760 and [ 18 F]F-Aza-MLN-4760 were evaluated with regard to the uptake in HEK-ACE2 and HEK-ACE cells and in vitro binding to tissue sections of HEK-ACE2 xenografts and normal organs of mice. Biodistribution and PET/CT imaging studies of [ 18 F]F-MLN-4760 and [ 18 F]F-Aza-MLN-4760 were performed using HEK-ACE2 and HEK-ACE xenografted nude mice. Crystallography data revealed an equal hACE2-binding mode for F-MLN-4760 as previously found for MLN-4760. Moreover, computer-based modeling indicated that similar binding to hACE2 and mACE2 holds true for both, F-MLN-4760 and F-Aza-MLN-4760, as is the case for MLN-4760. The IC 50 values were three-fold and seven-fold higher for F-MLN-4760 and F-Aza-MLN-4760, respectively, than for MLN-4760. [ 18 F]F-MLN-4760 and [ 18 F]F-Aza-MLN-4760 were obtained in 1.4 ± 0.3 GBq and 0.5 ± 0.1 GBq activity with > 99% radiochemical purity in a 5.3% and 1.2% radiochemical yield, respectively. Uptake in HEK-ACE2 cells was higher for [ 18 F]F-MLN-4760 (67 ± 9%) than for [ 18 F]F-Aza-MLN-4760 (37 ± 8%) after 3-h incubation while negligible uptake was seen in HEK-ACE cells (< 0.3%). [ 18 F]F-MLN-4760 and [ 18 F]F-Aza-MLN-4760 accumulated specifically in HEK-ACE2 xenografts of mice (13 ± 2% IA/g and 15 ± 2% IA/g at 1 h p.i.) with almost no uptake observed in HEK-ACE xenografts (< 0.3% IA/g). This was confirmed by PET/CT imaging, which also visualized unspecific accumulation in the gall bladder and intestinal tract. Both radiotracers showed specific and selective binding to ACE2 in vitro and in vivo. [ 18 F]F-MLN-4760 was, however, obtained in higher yields and the ACE2-binding affinity was superior over that of [ 18 F]F-Aza-MLN-4760. [ 18 F]F-MLN-4760 would, thus, be the candidate of choice for further development in view of its use for PET imaging of ACE2.
Organizational Affiliation:
Laboratory of Organic Chemistry, Department of Chemistry and Applied Biosciences, ETH Zurich, Zurich, 8093, Switzerland.