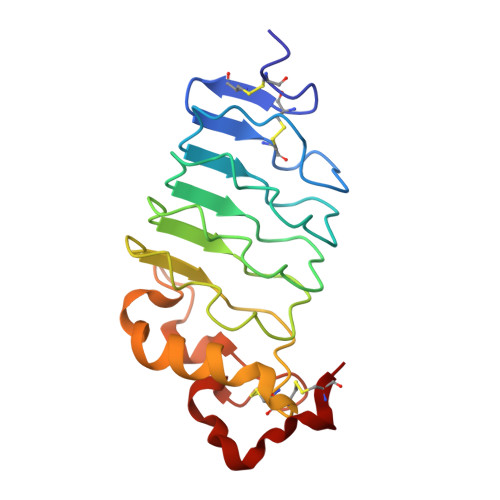
The High-Resolution Structure of a Variable Lymphocyte Receptor From Petromyzon marinus Capable of Binding to the Brain Extracellular Matrix.
Appelt, E.A., Thoden, J.B., Gehrke, S.A., Bachmeier, H.D., Rayment, I., Shusta, E.V., Holden, H.M.(2024) Proteins
- PubMed: 39601379
- DOI: https://doi.org/10.1002/prot.26768
- Primary Citation of Related Structures:
9CJ0 - PubMed Abstract:
Variable lymphocyte receptors (VLRs) are antigen receptors derived from the adaptive immune system of jawless vertebrates such as lamprey (Petromyzon marinus). First discovered in 2004, VLRs have been the subject of numerous biochemical and structural investigations. Due to their unique antigen binding properties, VLRs have been leveraged as possible drug delivery agents. One such VLR, previously identified and referred to as P1C10, was shown to bind to the brain extracellular matrix. Here, we present the high-resolution X-ray crystal structure of this VLR determined to 1.3 Å resolution. The fold is dominated by a six-stranded mixed β-sheet which provides a concave surface for possible antigen binding. Electron density corresponding to a 4-(2-hydroxyethyl)piperazine-1-propanesulfonic acid buffer molecule (HEPPS) was found in this region. By comparing the P1C10 molecular architecture and its buffer binding residues with those of other VLRs previously reported, it was possible to illustrate how this unique class of proteins can accommodate diverse binding partners. Additionally, we provide an analysis of the experimentally determined structure compared to the models generated by the commonly used AlphaFold and iTASSER structure prediction software packages.
Organizational Affiliation:
Department of Chemical and Biological Engineering, University of Wisconsin-Madison, Madison, Wisconsin, USA.