Does targeting Arg98 of FimH lead to high affinity antagonists?
Tomasic, T., Rabbani, S., Jakob, R.P., Reisner, A., Jakopin, Z., Maier, T., Ernst, B., Anderluh, M.(2020) Eur J Med Chem 211: 113093-113093
- PubMed: 33340913
- DOI: https://doi.org/10.1016/j.ejmech.2020.113093
- Primary Citation of Related Structures:
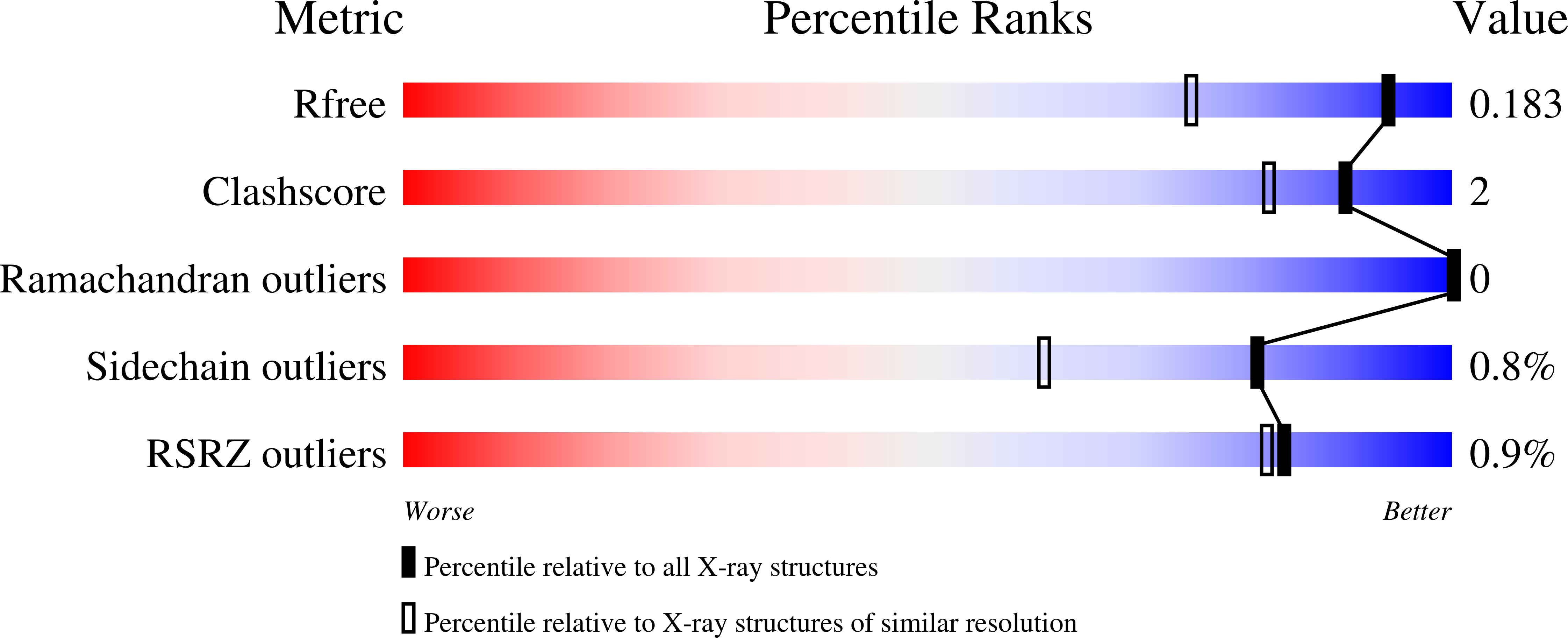
7AYN - PubMed Abstract:
Bacterial resistance has become an important challenge in the treatment of urinary tract infections. The underlying resistance mechanisms can most likely be circumvented with an antiadhesive approach, antagonizing the lectin FimH located at the tip of fimbriae of uropathogenic E. coli. Here we report on a novel series of FimH antagonists based on the 1-(α-d-mannopyranosyl)-4-phenyl-1,2,3-triazole scaffold, designed to incorporate carboxylic acid or ester functions to interact with FimH Arg98. The most potent representative of the series, ester 11e, displayed a K d value of 7.6 nM for the lectin domain of FimH with a general conclusion that all esters outperform carboxylates in terms of affinity. Surprisingly, all compounds from this new series exhibited improved binding affinities also for the R98A mutant, indicating another possible interaction contributing to binding. Our study on 1-(α-d-mannopyranosyl)-4-phenyl-1,2,3-triazole-based FimH antagonists offers proof that targeting Arg98 side chain by a "chemical common sense", i.e. by introduction of the acidic moiety to form ionic bond with Arg98 is most likely unsuitable approach to boost FimH antagonists' potency.
Organizational Affiliation:
University of Ljubljana, Faculty of Pharmacy, Aškerčeva 7, 1000, Ljubljana, Slovenia.