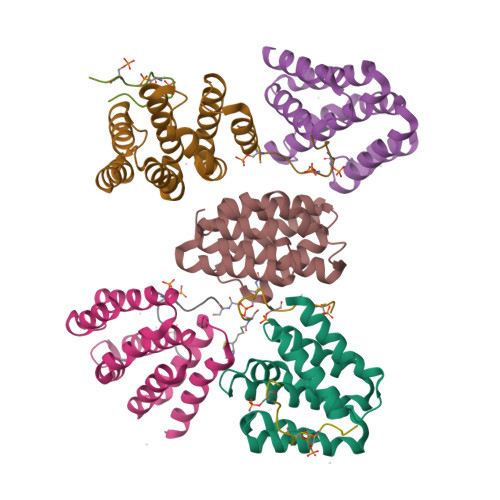
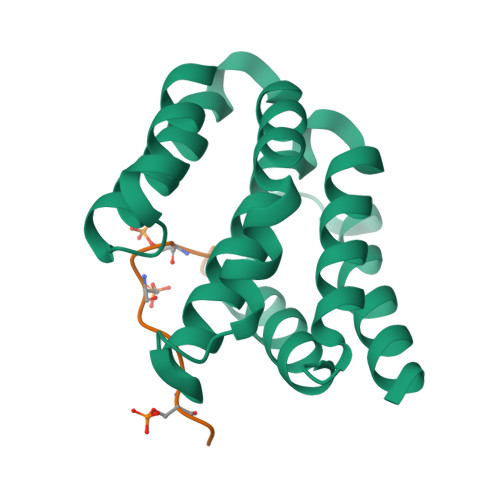
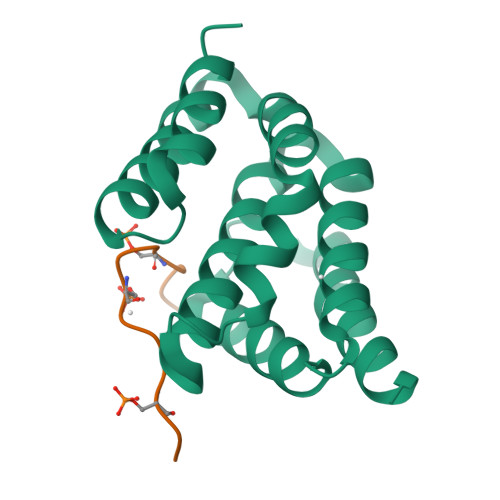
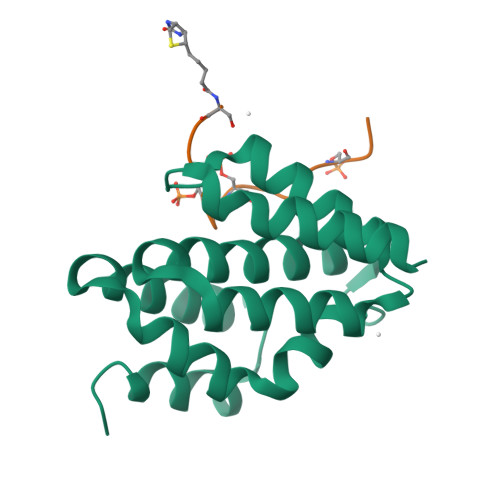
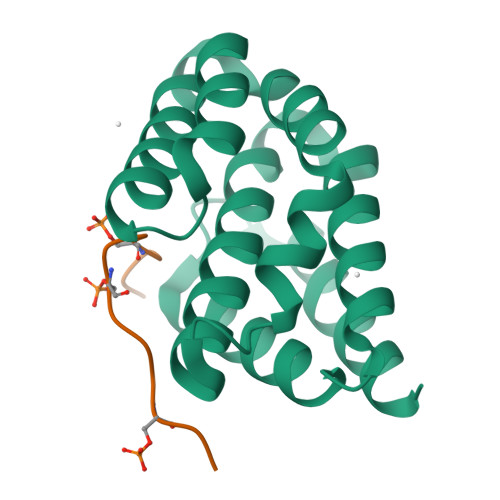
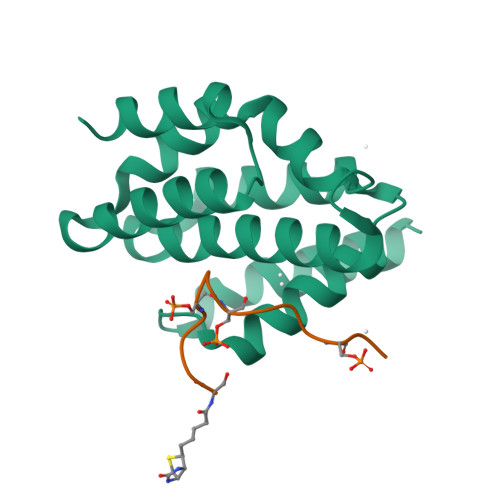
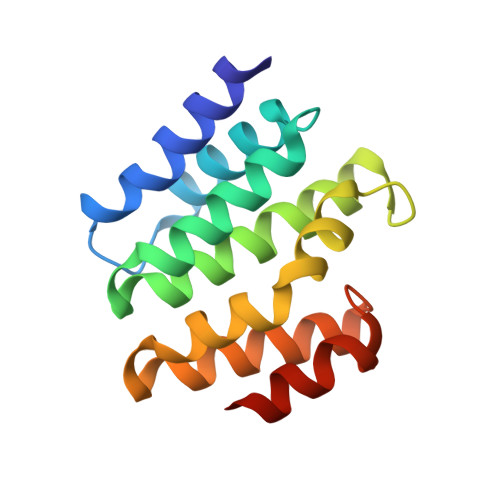
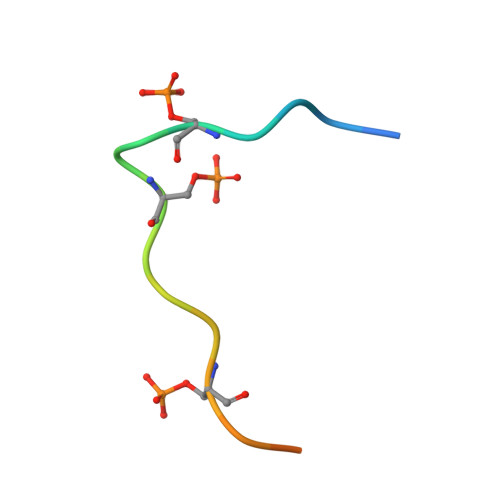
Structural basis for the recognition of the S2, S5-phosphorylated RNA polymerase II CTD by the mRNA anti-terminator protein hSCAF4.
Zhou, M., Ehsan, F., Gan, L., Dong, A., Li, Y., Liu, K., Min, J.(2022) FEBS Lett 596: 249-259
- PubMed: 34897689
- DOI: https://doi.org/10.1002/1873-3468.14256
- Primary Citation of Related Structures:
6XKB - PubMed Abstract:
The C-terminal domain (CTD) of RNA polymerase II serves as a binding platform for numerous enzymes and transcription factors involved in nascent RNA processing and the transcription cycle. The S2, S5-phosphorylated CTD is recognized by the transcription factor SCAF4, which functions as a transcription anti-terminator by preventing early mRNA transcript cleavage and polyadenylation. Here, we measured the binding affinities of differently modified CTD peptides by hSCAF4 and solved the complex structure of the hSCAF4-CTD-interaction domain (CID) bound to a S2, S5-quadra-phosphorylated CTD peptide. Our results revealed that the S2, S5-quadra-phosphorylated CTD peptide adopts a trans conformation and is located in a positively charged binding groove of hSCAF4-CID. Although hSCAF4-CID has almost the same binding pattern to the CTD as other CID-containing proteins, it preferentially binds to the S2, S5-phosphorylated CTD. Our findings provide insight into the regulatory mechanism of hSCAF4 in transcription termination.
Organizational Affiliation:
Hubei Key Laboratory of Genetic Regulation and Integrative Biology, School of Life Sciences, Central China Normal University, Wuhan, China.