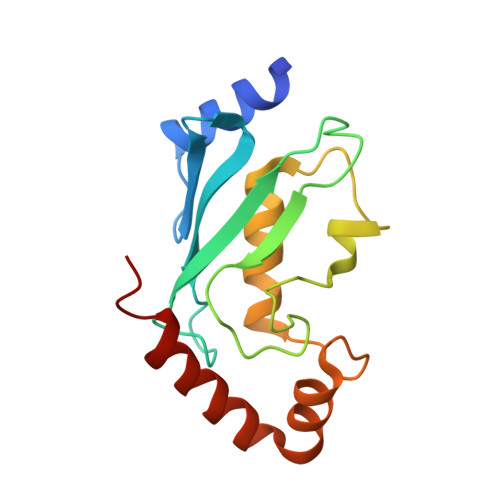
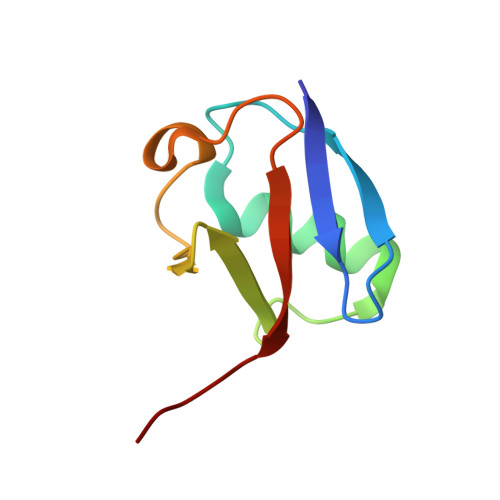
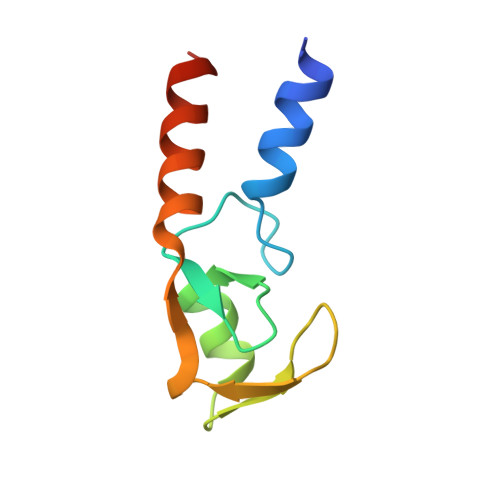
A tri-ionic anchor mechanism drives Ube2N-specific recruitment and K63-chain ubiquitination in TRIM ligases.
Kiss, L., Zeng, J., Dickson, C.F., Mallery, D.L., Yang, J.C., McLaughlin, S.H., Boland, A., Neuhaus, D., James, L.C.(2019) Nat Commun 10: 4502-4502
- PubMed: 31582740
- DOI: https://doi.org/10.1038/s41467-019-12388-y
- Primary Citation of Related Structures:
6S53 - PubMed Abstract:
The cytosolic antibody receptor TRIM21 possesses unique ubiquitination activity that drives broad-spectrum anti-pathogen targeting and underpins the protein depletion technology Trim-Away. This activity is dependent on formation of self-anchored, K63-linked ubiquitin chains by the heterodimeric E2 enzyme Ube2N/Ube2V2. Here we reveal how TRIM21 facilitates ubiquitin transfer and differentiates this E2 from other closely related enzymes. A tri-ionic motif provides optimally distributed anchor points that allow TRIM21 to wrap an Ube2N~Ub complex around its RING domain, locking the closed conformation and promoting ubiquitin discharge. Mutation of these anchor points inhibits ubiquitination with Ube2N/Ube2V2, viral neutralization and immune signalling. We show that the same mechanism is employed by the anti-HIV restriction factor TRIM5 and identify spatially conserved ionic anchor points in other Ube2N-recruiting RING E3s. The tri-ionic motif is exclusively required for Ube2N but not Ube2D1 activity and provides a generic E2-specific catalysis mechanism for RING E3s.
Organizational Affiliation:
Medical Research Council Laboratory of Molecular Biology, Cambridge, UK.