Lactate Racemase Nickel-Pincer Cofactor Operates by a Proton-Coupled Hydride Transfer Mechanism.
Rankin, J.A., Mauban, R.C., Fellner, M., Desguin, B., McCracken, J., Hu, J., Varganov, S.A., Hausinger, R.P.(2018) Biochemistry 57: 3244-3251
- PubMed: 29489337
- DOI: https://doi.org/10.1021/acs.biochem.8b00100
- Primary Citation of Related Structures:
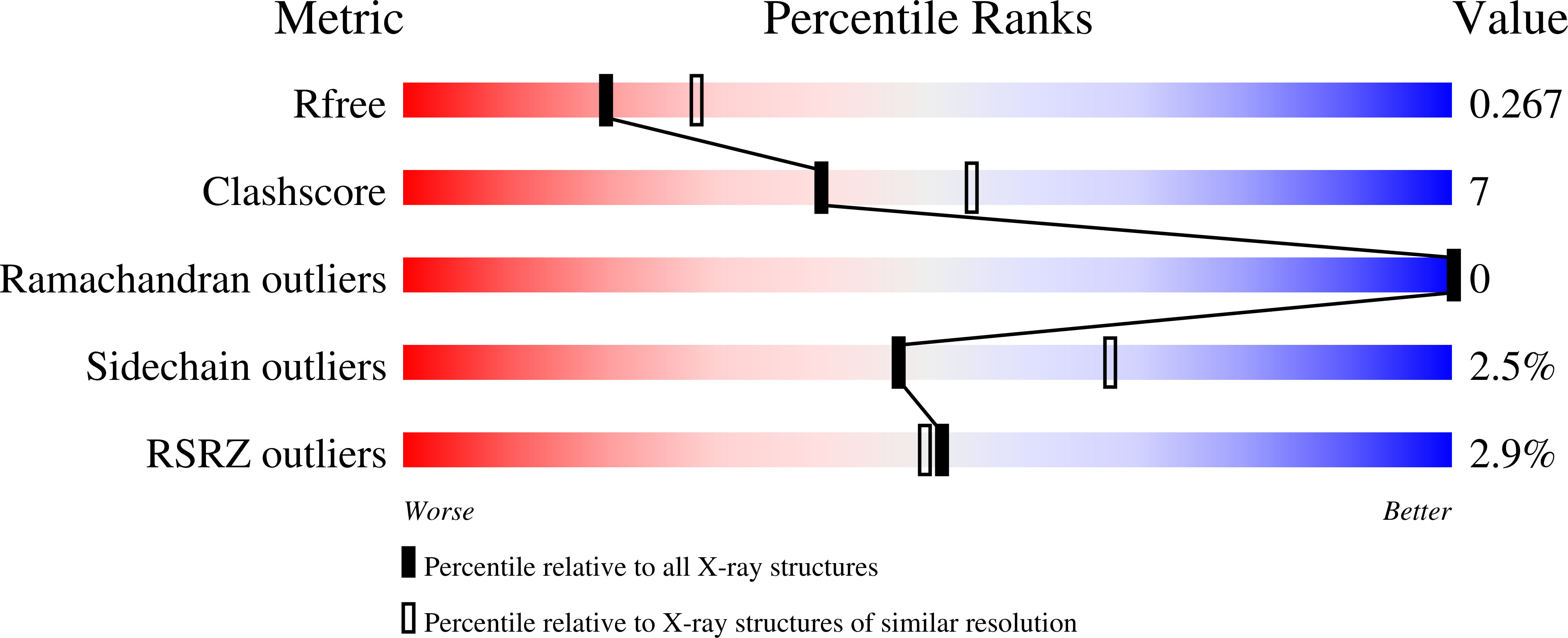
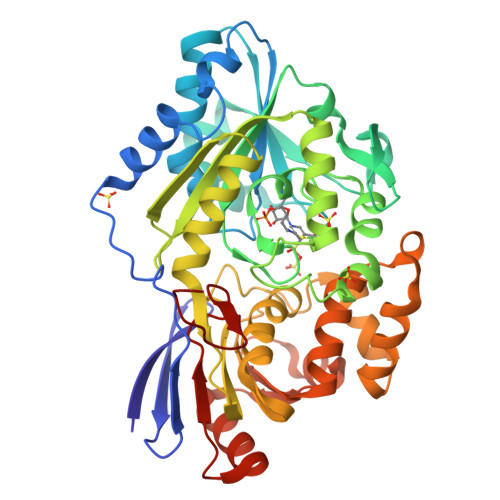
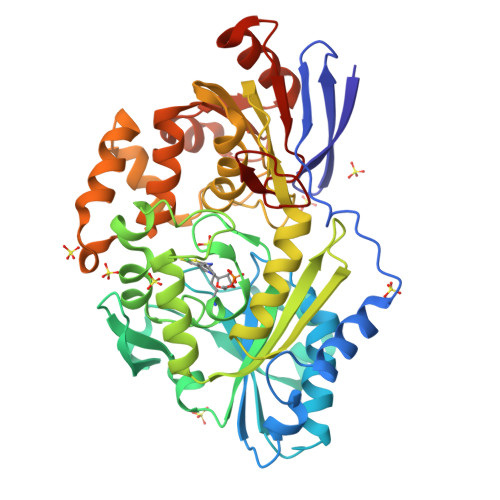
6C1W - PubMed Abstract:
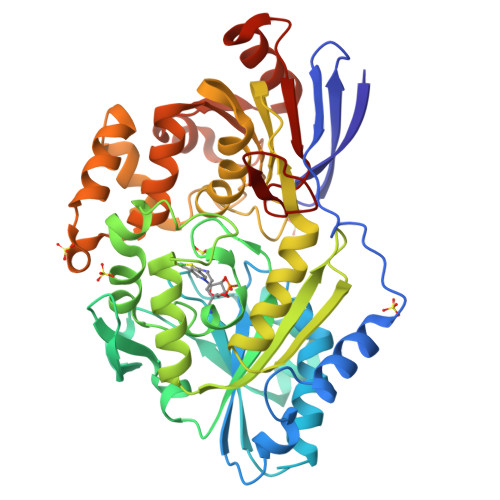
Lactate racemase (LarA) of Lactobacillus plantarum contains a novel organometallic cofactor with nickel coordinated to a covalently tethered pincer ligand, pyridinium-3-thioamide-5-thiocarboxylic acid mononucleotide, but its function in the enzyme mechanism has not been elucidated. This study presents direct evidence that the nickel-pincer cofactor facilitates a proton-coupled hydride transfer (PCHT) mechanism during LarA-catalyzed lactate racemization. No signal was detected by electron paramagnetic resonance spectroscopy for LarA in the absence or presence of substrate, consistent with a +2 metal oxidation state and inconsistent with a previously proposed proton-coupled electron transfer mechanism. Pyruvate, the predicted intermediate for a PCHT mechanism, was observed in quenched solutions of LarA. A normal substrate kinetic isotope effect ( k H / k D of 3.11 ± 0.17) was established using 2-α- 2 H-lactate, further supporting a PCHT mechanism. UV-visible spectroscopy revealed a lactate-induced perturbation of the cofactor spectrum, notably increasing the absorbance at 340 nm, and demonstrated an interaction of the cofactor with the inhibitor sulfite. A crystal structure of LarA provided greater resolution (2.4 Å) than previously reported and revealed sulfite binding to the pyridinium C4 atom of the reduced pincer cofactor, mimicking hydride reduction during a PCHT catalytic cycle. Finally, computational modeling supports hydride transfer to the cofactor at the C4 position or to the nickel atom, but with formation of a nickel-hydride species requiring dissociation of the His200 metal ligand. In aggregate, these studies provide compelling evidence that the nickel-pincer cofactor acts by a PCHT mechanism.
Organizational Affiliation:
Department of Chemistry , University of Nevada , Reno , Nevada 89557 , United States.