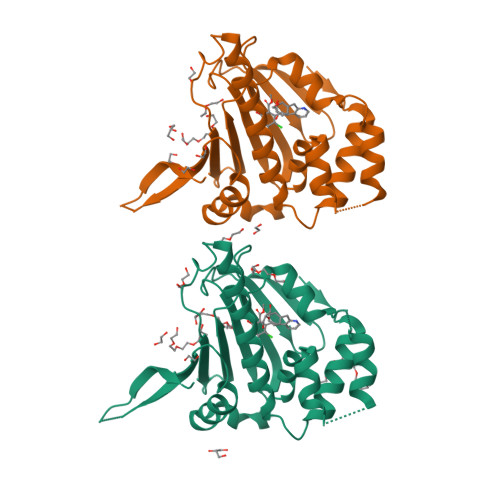
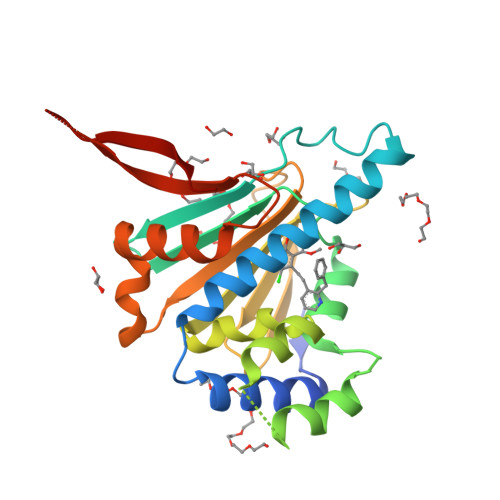
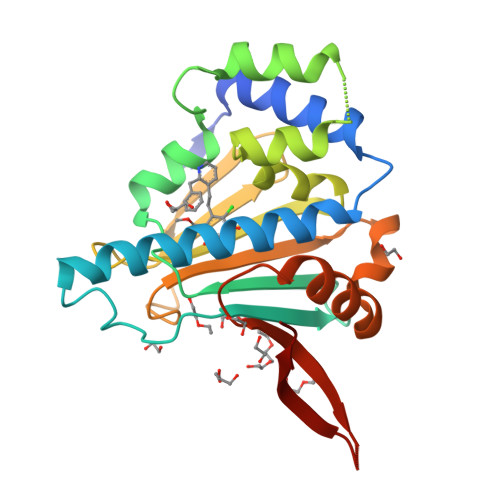
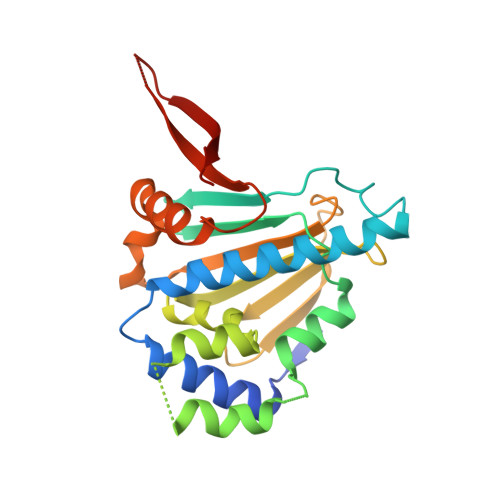
Trifunctional High-Throughput Screen Identifies Promising Scaffold To Inhibit Grp94 and Treat Myocilin-Associated Glaucoma.
Huard, D.J.E., Crowley, V.M., Du, Y., Cordova, R.A., Sun, Z., Tomlin, M.O., Dickey, C.A., Koren, J., Blair, L., Fu, H., Blagg, B.S.J., Lieberman, R.L.(2018) ACS Chem Biol 13: 933-941
- PubMed: 29402077
- DOI: https://doi.org/10.1021/acschembio.7b01083
- Primary Citation of Related Structures:
6ASP, 6ASQ - PubMed Abstract:
Gain-of-function mutations within the olfactomedin (OLF) domain of myocilin result in its toxic intracellular accumulation and hasten the onset of open-angle glaucoma. The absence of myocilin does not cause disease; therefore, strategies aimed at eliminating myocilin could lead to a successful glaucoma treatment. The endoplasmic reticulum Hsp90 paralog Grp94 accelerates OLF aggregation. Knockdown or pharmacological inhibition of Grp94 in cells facilitates clearance of mutant myocilin via a non-proteasomal pathway. Here, we expanded our support for targeting Grp94 over cytosolic paralogs Hsp90α and Hsp90β. We then developed a high-throughput screening assay to identify new chemical matter capable of disrupting the Grp94/OLF interaction. When applied to a blind, focused library of 17 Hsp90 inhibitors, our miniaturized single-read in vitro thioflavin T -based kinetics aggregation assay exclusively identified compounds that target the chaperone N-terminal nucleotide binding site. In follow up studies, one compound (2) decreased the extent of co-aggregation of Grp94 with OLF in a dose-dependent manner in vitro, and enabled clearance of the aggregation-prone full-length myocilin variant I477N in cells without inducing the heat shock response or causing cytotoxicity. Comparison of the co-crystal structure of compound 2 and another non-selective hit in complex with the N-terminal domain of Grp94 reveals a docking mode tailored to Grp94 and explains its selectivity. A new lead compound has been identified, supporting a targeted chemical biology assay approach to develop a protein degradation-based therapy for myocilin-associated glaucoma by selectively inhibiting Grp94.
Organizational Affiliation:
School of Chemistry & Biochemistry , Georgia Institute of Technology , Atlanta , Georgia 30332 , United States.