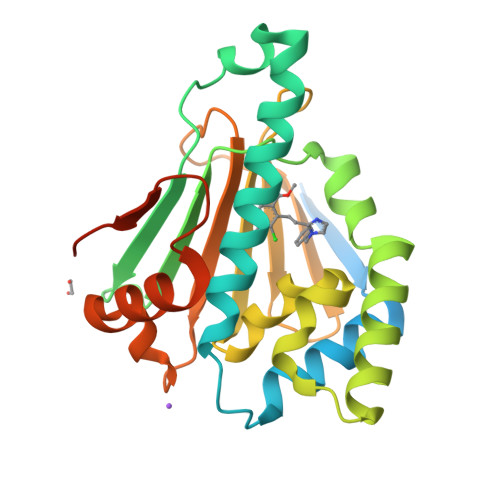
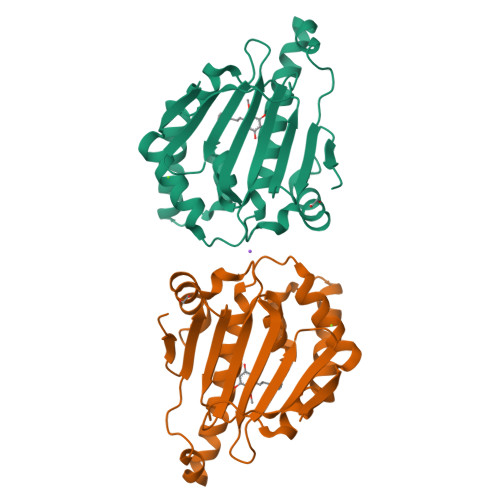
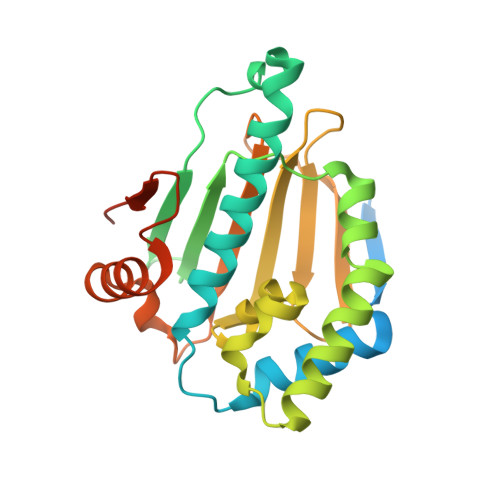
Structure Based Design of a Grp94-Selective Inhibitor: Exploiting a Key Residue in Grp94 To Optimize Paralog-Selective Binding.
Que, N.L.S., Crowley, V.M., Duerfeldt, A.S., Zhao, J., Kent, C.N., Blagg, B.S.J., Gewirth, D.T.(2018) J Med Chem 61: 2793-2805
- PubMed: 29528635
- DOI: https://doi.org/10.1021/acs.jmedchem.7b01608
- Primary Citation of Related Structures:
5VYY, 5WMT, 6BAW, 6C91, 6CEO - PubMed Abstract:
Grp94 and Hsp90, the ER and cytoplasmic hsp90 paralogs, share a conserved ATP-binding pocket that has been targeted for therapeutics. Paralog-selective inhibitors may lead to drugs with fewer side effects. Here, we analyzed 1 (BnIm), a benzyl imidazole resorcinylic inhibitor, for its mode of binding. The structures of 1 bound to Hsp90 and Grp94 reveal large conformational changes in Grp94 but not Hsp90 that expose site 2, a binding pocket adjacent to the central ATP cavity that is ordinarily blocked. The Grp94:1 structure reveals a flipped pose of the resorcinylic scaffold that inserts into the exposed site 2. We exploited this flipped binding pose to develop a Grp94-selective derivative of 1. Our structural analysis shows that the ability of the ligand to insert its benzyl imidazole substituent into site 1, a different side pocket off the ATP binding cavity, is the key to exposing site 2 in Grp94.
Organizational Affiliation:
Hauptman-Woodward Medical Research Institute , Buffalo , New York 14203 , United States.