TLR4/MD-2 activation by a synthetic agonist with no similarity to LPS.
Wang, Y., Su, L., Morin, M.D., Jones, B.T., Whitby, L.R., Surakattula, M.M., Huang, H., Shi, H., Choi, J.H., Wang, K.W., Moresco, E.M., Berger, M., Zhan, X., Zhang, H., Boger, D.L., Beutler, B.(2016) Proc Natl Acad Sci U S A 113: E884-E893
- PubMed: 26831104
- DOI: https://doi.org/10.1073/pnas.1525639113
- Primary Citation of Related Structures:
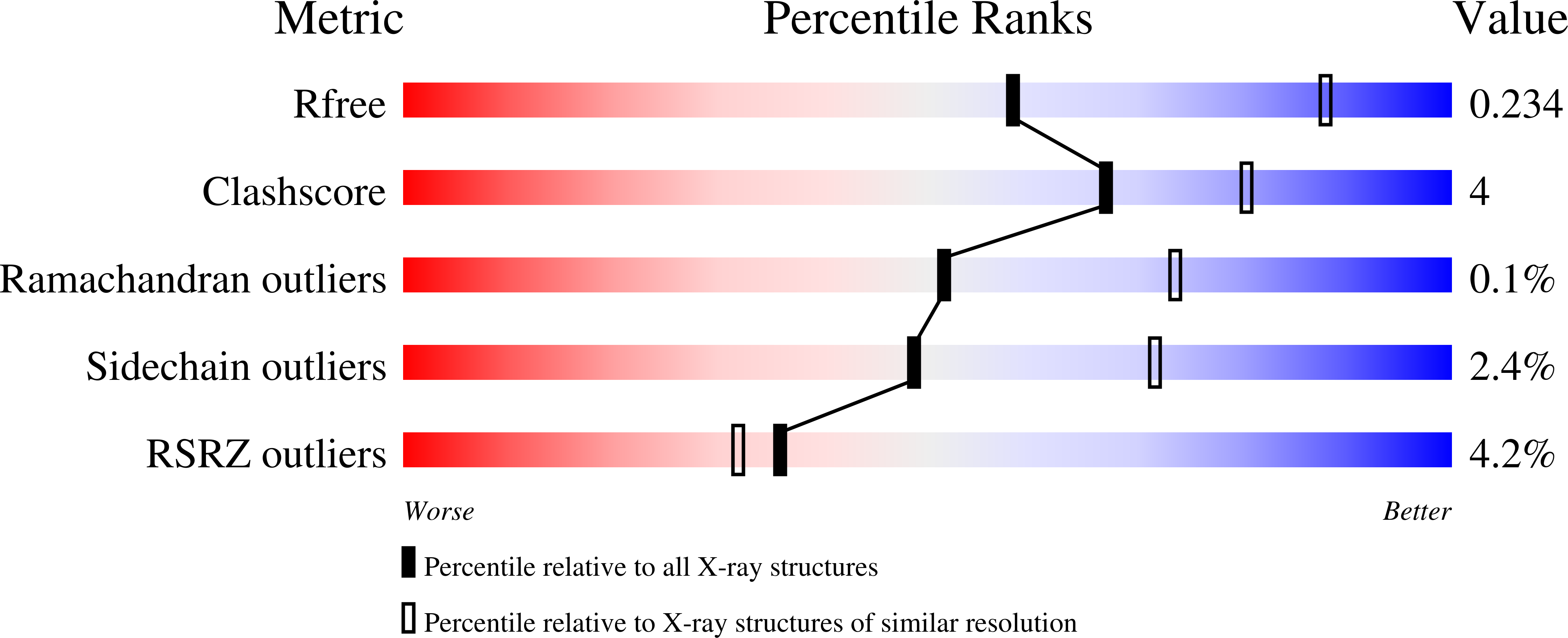
5IJB, 5IJC, 5IJD - PubMed Abstract:
Structurally disparate molecules reportedly engage and activate Toll-like receptor (TLR) 4 and other TLRs, yet the interactions that mediate binding and activation by dissimilar ligands remain unknown. We describe Neoseptins, chemically synthesized peptidomimetics that bear no structural similarity to the established TLR4 ligand, lipopolysaccharide (LPS), but productively engage the mouse TLR4 (mTLR4)/myeloid differentiation factor 2 (MD-2) complex. Neoseptin-3 activates mTLR4/MD-2 independently of CD14 and triggers canonical myeloid differentiation primary response gene 88 (MyD88)- and Toll-interleukin 1 receptor (TIR) domain-containing adaptor inducing IFN-beta (TRIF)-dependent signaling. The crystal structure mTLR4/MD-2/Neoseptin-3 at 2.57-Å resolution reveals that Neoseptin-3 binds as an asymmetrical dimer within the hydrophobic pocket of MD-2, inducing an active receptor complex similar to that induced by lipid A. However, Neoseptin-3 and lipid A form dissimilar molecular contacts to achieve receptor activation; hence strong TLR4/MD-2 agonists need not mimic LPS.
Organizational Affiliation:
Center for the Genetics of Host Defense, University of Texas Southwestern Medical Center, Dallas, TX 75390;