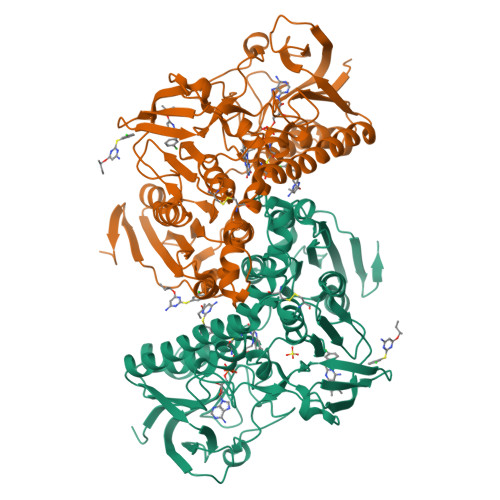
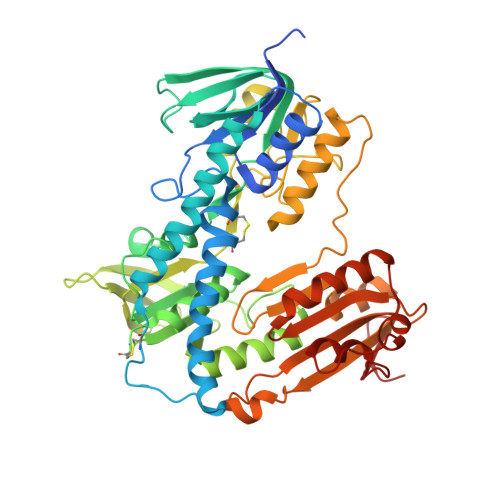
Inhibition of Leishmania infantum trypanothione reductase by diaryl sulfide derivatives.
Saccoliti, F., Angiulli, G., Pupo, G., Pescatori, L., Madia, V.N., Messore, A., Colotti, G., Fiorillo, A., Scipione, L., Gramiccia, M., Di Muccio, T., Di Santo, R., Costi, R., Ilari, A.(2017) J Enzyme Inhib Med Chem 32: 304-310
- PubMed: 28098499
- DOI: https://doi.org/10.1080/14756366.2016.1250755
- Primary Citation of Related Structures:
5EBK - PubMed Abstract:
The study presented here aimed at identifying a new class of compounds acting against Leishmania parasites, the causative agent of Leishmaniasis. For this purpose, the thioether derivatives of our in-house library have been evaluated in whole-cell screening assays in order to determine their in vitro activity against Leishmania protozoan. Among them, promising results have been achieved with compound RDS 777 (6-(sec-butoxy)-2-((3-chlorophenyl)thio)pyrimidin-4-amine) (IC 50 = 29.43 µM), which is able to impair the mechanism of the parasite defence against the reactive oxygen species by inhibiting the trypanothione reductase (TR) with high efficiency (K i 0.25 ± 0.18 µM). The X-ray structure of L. infantum TR in complex with RDS 777 disclosed the mechanism of action of this compound that binds to the catalytic site and engages in hydrogen bonds the residues more involved in the catalysis, namely Glu466', Cys57 and Cys52, thereby inhibiting the trypanothione binding and avoiding its reduction.
Organizational Affiliation:
a Istituto Pasteur-Fondazione Cenci Bolognetti , Dipartimento di Chimica e Tecnologie del Farmaco , "Sapienza" Università di Roma , Roma , Italia.