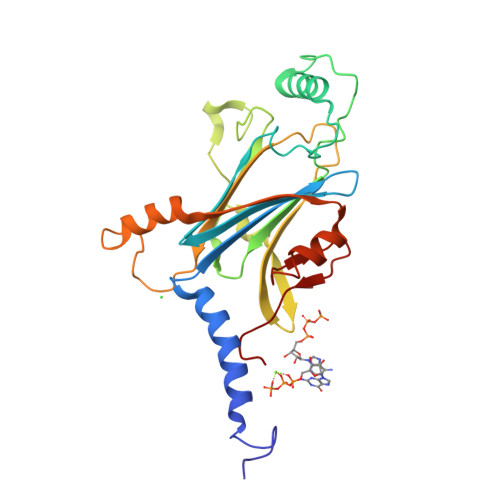
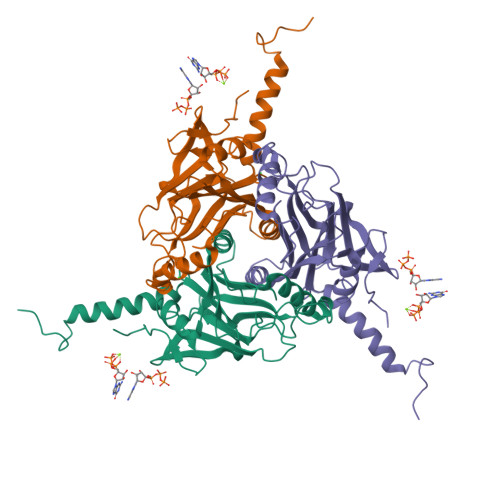
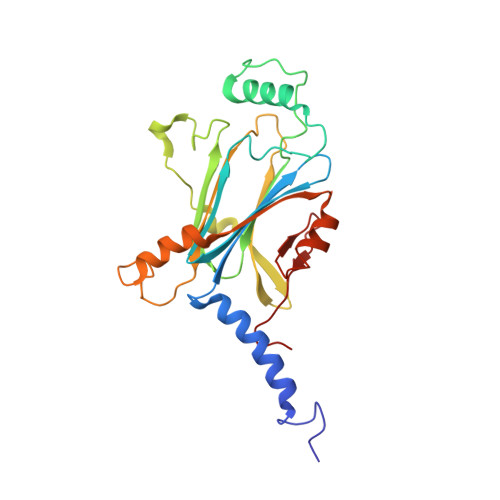
In cellulo structure determination of a novel cypovirus polyhedrin.
Axford, D., Ji, X., Stuart, D.I., Sutton, G.(2014) Acta Crystallogr D Biol Crystallogr 70: 1435-1441
- PubMed: 24816111
- DOI: https://doi.org/10.1107/S1399004714004714
- Primary Citation of Related Structures:
4OTS, 4OTV - PubMed Abstract:
This work demonstrates that with the use of a microfocus synchrotron beam the structure of a novel viral polyhedrin could be successfully determined from microcrystals within cells, removing the preparatory step of sample isolation and maintaining a favourable biological environment. The data obtained are of high quality, comparable to that obtained from isolated crystals, and enabled a facile structure determination. A small but significant difference is observed between the unit-cell parameters and the mosaic spread of in cellulo and isolated crystals, suggesting that even these robust crystals are adversely affected by removal from the cell.
Organizational Affiliation:
Diamond Light Source Ltd, Harwell Oxford, Didcot OX11 0DE, England.