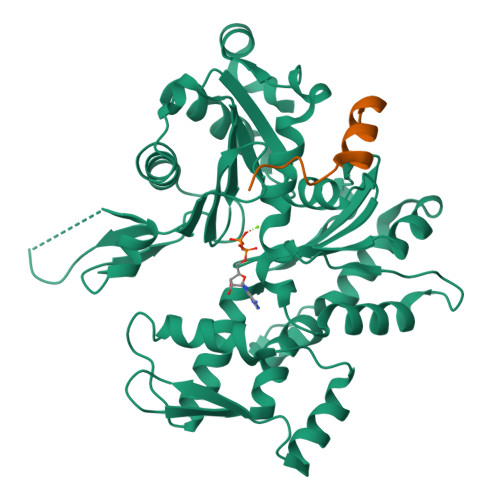
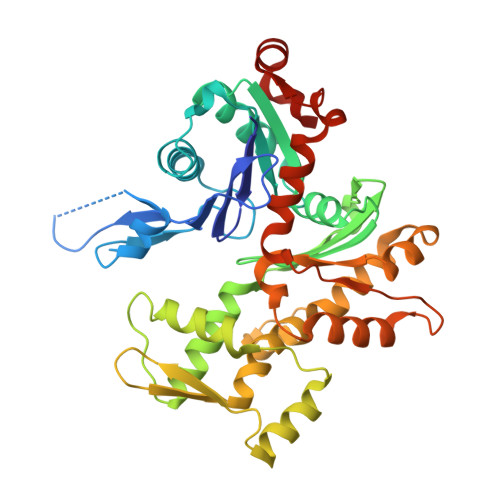
How a single residue in individual beta-thymosin/WH2 domains controls their functions in actin assembly
Didry, D., Cantrelle, F.X., Husson, C., Roblin, P., Moorthy, A.M., Perez, J., Le Clainche, C., Hertzog, M., Guittet, E., Carlier, M.F., van Heijenoort, C., Renault, L.(2012) EMBO J 31: 1000-1013
- PubMed: 22193718
- DOI: https://doi.org/10.1038/emboj.2011.461
- Primary Citation of Related Structures:
3SJH, 3U8X, 3U9D, 3U9Z - PubMed Abstract:
β-Thymosin (βT) and WH2 domains are widespread, intrinsically disordered actin-binding peptides that display significant sequence variability and different regulations of actin self-assembly in motile and morphogenetic processes. Here, we reveal the structural mechanisms by which, in their 1:1 stoichiometric complexes with actin, they either inhibit assembly by sequestering actin monomers like Thymosin-β4, or enhance motility by directing polarized filament assembly like Ciboulot βT. We combined mutational, functional or structural analysis by X-ray crystallography, SAXS (small angle X-ray scattering) and NMR on Thymosin-β4, Ciboulot, TetraThymosinβ and the long WH2 domain of WASP-interacting protein. The latter sequesters G-actin with the same molecular mechanisms as Thymosin-β4. Functionally different βT/WH2 domains differ by distinct dynamics of their C-terminal half interactions with G-actin pointed face. These C-terminal interaction dynamics are controlled by the strength of electrostatic interactions with G-actin. At physiological ionic strength, a single salt bridge with actin located next to their central LKKT/V motif induces G-actin sequestration in both isolated long βT and WH2 domains. The results open perspectives for elucidating the functions of βT/WH2 domains in other modular proteins.
Organizational Affiliation:
Laboratoire d'Enzymologie et Biochimie Structurales, Centre de Recherche de Gif, CNRS, Gif-sur-Yvette, France.