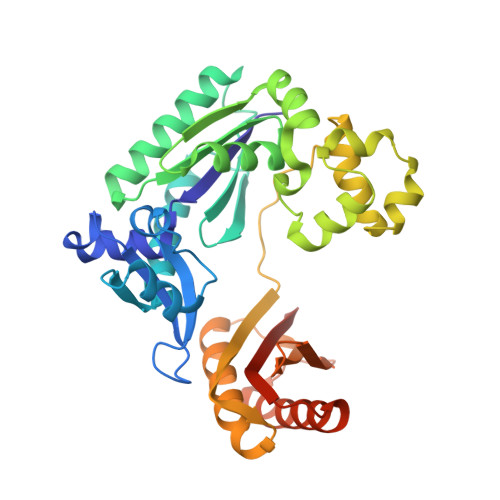
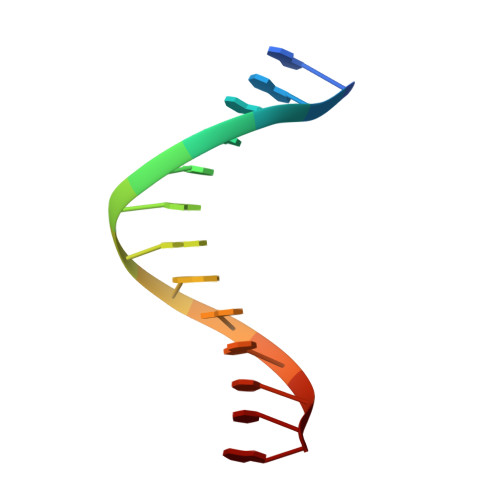
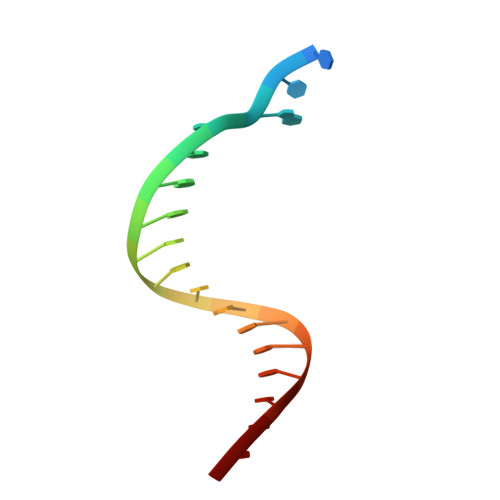
Dpo4 is hindered in extending a G.T mismatch by a reverse wobble
Trincao, J., Johnson, R.E., Wolfle, W.T., Escalante, C.R., Prakash, S., Prakash, L., Aggarwal, A.K.(2004) Nat Struct Mol Biol 11: 457-462
- PubMed: 15077104
- DOI: https://doi.org/10.1038/nsmb755
- Primary Citation of Related Structures:
1S97, 1S9F - PubMed Abstract:
The ability or inability of a DNA polymerase to extend a mispair directly affects the establishment of genomic mutations. We report here kinetic analyses of the ability of Dpo4, a Y-family polymerase from Sulfolobus solfataricus, to extend from all mispairs opposite a template G or T. Dpo4 is equally inefficient at extending these mispairs, which include, surprisingly, a G.T mispair expected to conform closely to Watson-Crick geometry. To elucidate the basis of this, we solved the structure of Dpo4 bound to G.T-mispaired primer template in the presence of an incoming nucleotide. As a control, we also determined the structure of Dpo4 bound to a matched A-T base pair at the primer terminus. The structures offer a basis for the low efficiency of Dpo4 in extending a G.T mispair: a reverse wobble that deflects the primer 3'-OH away from the incoming nucleotide.
Organizational Affiliation:
Structural Biology Program, Department of Physiology and Biophysics, Mount Sinai School of Medicine, Box 1677, 1425 Madison Avenue, New York, New York 10029, USA.