Full Text |
QUERY: Gene Name = "nagH" | MyPDB Login | Search API |
| Search Summary | This query matches 6 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameSymmetry TypeSCOP Classification | 1 to 6 of 6 Structures Page 1 of 1 Sort by
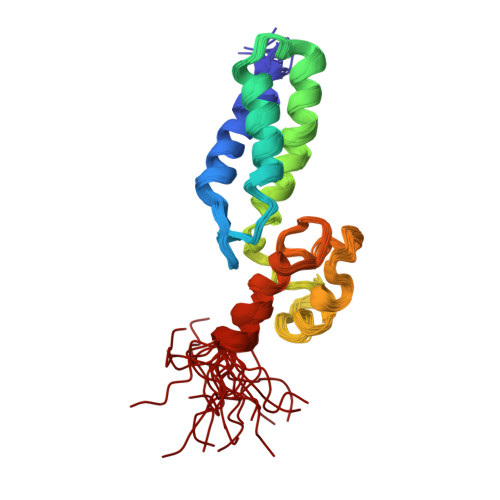
Solution structure of a dockerin-containing modular pair from a family 84 glycoside hydrolaseChitayat, S., Adams, J.J., Bayer, E.A., Smith, S.P. (2008) J Mol Biology 381: 1202-1212
Solution NMR Structure of a Non-canonical galactose-binding CBM32 from Clostridium perfringensGrondin, J.M., Chitayat, S., Ficko-Blean, E., Boraston, A.B., Smith, S.P. (2014) J Mol Biology 426: 869-880
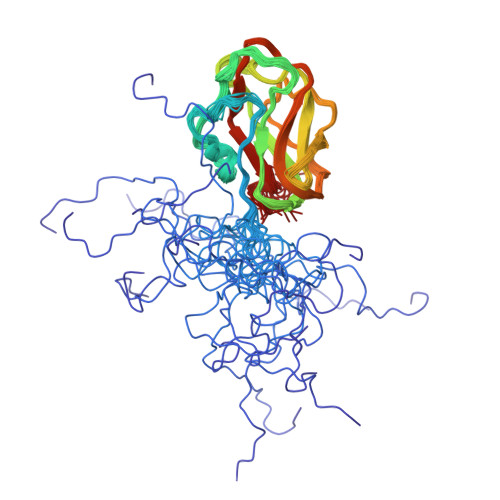
The Cohesin-Dockerin Complex of NagJ and NagH from Clostridium perfringensAdams, J.J., Boraston, A., Smith, S.P. (2008) Proc Natl Acad Sci U S A 105: 12194-12199
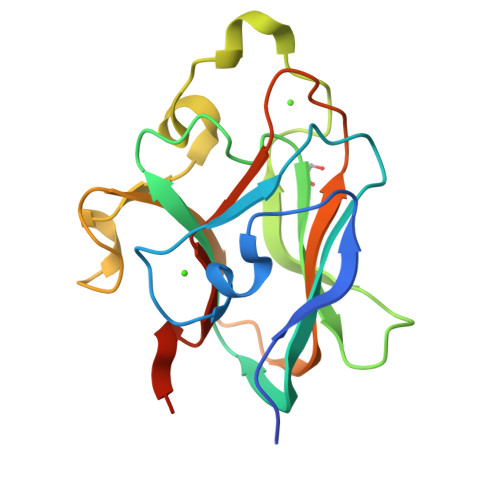
Crystal structure of CBM32-4 from the Clostridium perfringens NagHGrondin, J.M., Ficko-Blean, E., Boraston, A.B., Smith, S.P. To be published
Structure of CpGH84A(2019) Glycobiology 30: 49-57
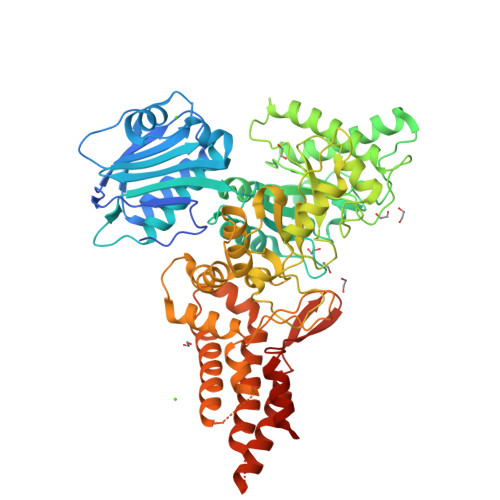
Crystal structure of salicylate 5-hydroxylase NagGH (a Rieske non-heme iron-dependent monooxgenase)(2021) Appl Environ Microbiol 87:
1 to 6 of 6 Structures Page 1 of 1 Sort by |